Submitted:
02 September 2024
Posted:
02 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
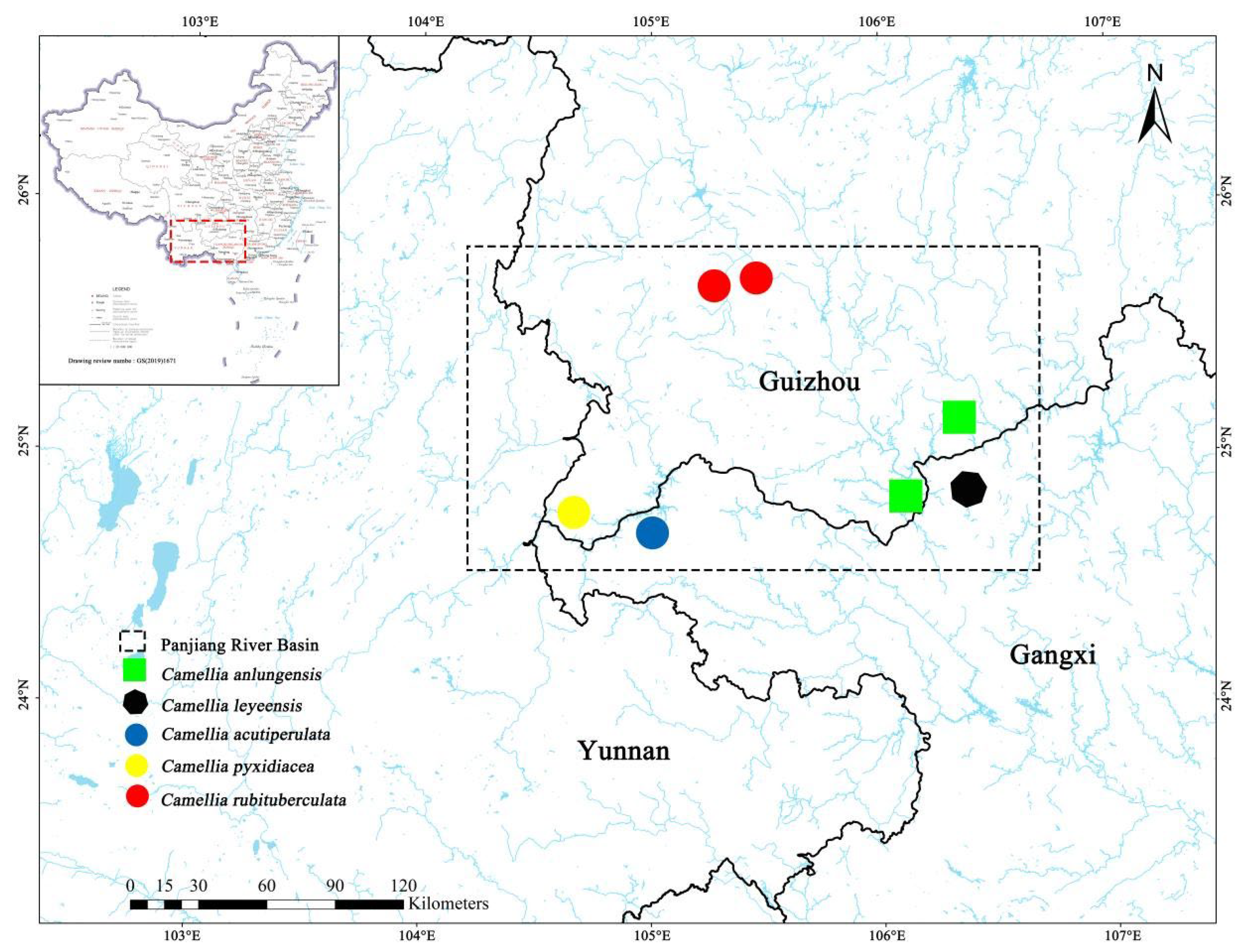
2. Materials and Methods
| Species name | Location | Specimen number | Elevation(m) | Longitude and latitude |
|---|---|---|---|---|
| C. anlungensis | Wangmo County, Guizhou, China | GZAC, LZ20211204 | 449 | 106°16′40.69″ E, 25°2′51.65″ N |
| C. leyeensis | Leye County, Guangxi, China | GZAC, LZ20210413 | 684 | 106°17′31.48″ E, 24°51′29.98″ N |
| C. acutiperulata | Longlin County, Guangxi, China | GZAC, LZ20221103 | 851 | 104°52′39.66″ E, 24°39′31.59″ N |
| C. pyxidiacea | Xingyi City, Guizhou, China | GZAC, LZ20211204 | 837 | 104°32′18.18″ E, 24°47′53.12″ N |
| C. rubituberculata | Xingren County, Guizhou, China | GZAC, LZ20210411 | 1297 | 105°21′08.10″ E, 25°43′50.08″ N |
2.2. Experimental Research Method
2.2.1. Data Statistics
2.2.2. Micromorphological Characteristics of Pollen
2.2.3. Chloroplast DNA Acquisition and Genome Assembly
2.2.4. Expansion and Contraction of the IR Boundary
2.2.5. Phylogenetic Tree
3.1. Morphological Studies
| Species name | C.anlungensis | C.leyeensis | C.acutiperulata | pyxidiacea | C.rubituberculata |
|---|---|---|---|---|---|
| Leaf type | leaves thin, leathery, obovate | leaves thin, leathery, obovate | leaves thin, coriaceous, elliptic | leaves thin, coriaceous, elliptic | leaves thin coriaceous, elliptic |
| - length (cm) | 10.72 ± 1.05 | 10.73 ± 0.55 | 12.37 ± 1.09 | 10.88 ± 1.53 | 10.68 ± 0.46 |
| - width (cm) | 3.44 ± 0.55 | 3.32 ± 0.38 | 3.92 ± 0.48 | 4.54 ± 0.30 | 4.08 ± 0.49 |
| Flower type | obovate | obovate | elliptic | elliptic | elliptic |
| - color | white | white | white | with pink petal apexes | red |
| - length (cm) | 2.42 ± 0.26 | 2.55 ± 0.27 | 3.71 ± 0.18 | 2.66 ± 0.14 | 2.98 ± 0.17 |
| - width (cm) | 1.3 ± 0.17 | 1.42 ± 0.24 | 1.62 ± 0.16 | 1.33 ± 0.2 | 2.71 ± 0.39 |
| Number of petals | 11 - 13 | 10 - 12 | 9 - 12 | 6 - 9 | 6 - 8 |
| Sepal | ovate | ovate | ovate, apex pointed, margin membranous | ovate | semicircular |
| Number of calyces | 5 - 7 | 5 - 8 | 5 - 7 | 6 - 8 | 7 - 9 |
| Fruit shape | capsule verrucose, subglobose, epidermis with verrucose projections | capsule verrucose, subglobose, epidermis with verrucose projections | capsule verrucose, subglobose, testa transversely lobed | fruit compressed globose, skin with verrucose protuberance and obvious parting | capsule verrucose, globose, seed coat shallowly undulate |
| - diameter (cm) | 1.75 ± 0.23 | 1.63 ± 0.17 | 1.70 ± 0.29 | 3.29 ± 0.40 | 4.99 ± 0.11 |
| Shell thickness (mm) | 1.79 ± 0.18 | 1.72± 0.34 | 3.49 ± 0.61 | 5.06 ± 1.29 | 13.16 ± 0.7 |

3.2. Micromorphological Characteristics of Pollen
| Species | Pollen Shape | Polar view | Equatorial view | Pollen aperture | Pollen wall ornamentation | Ridge | |
|---|---|---|---|---|---|---|---|
| C. anlungensis | oblate spherical | Triangular | Oval | Colpus | Uneven | Wavy | |
| C. leyeensis | oblate spherical | Triangular | Oval | Colpus | Uneven | Wavy | |
| C. acutiperulata | oblate spherical | Triangular | Oval | Colpus | Uneven | Wavy | |
| C. pyxidiacea | oblate spherical | Triangular | Oval | Colpus | Uneven | Wavy | |
| C. rubituberculata | oblate spherical | Oval | Oval | Colpus | Uneven | Wavy |
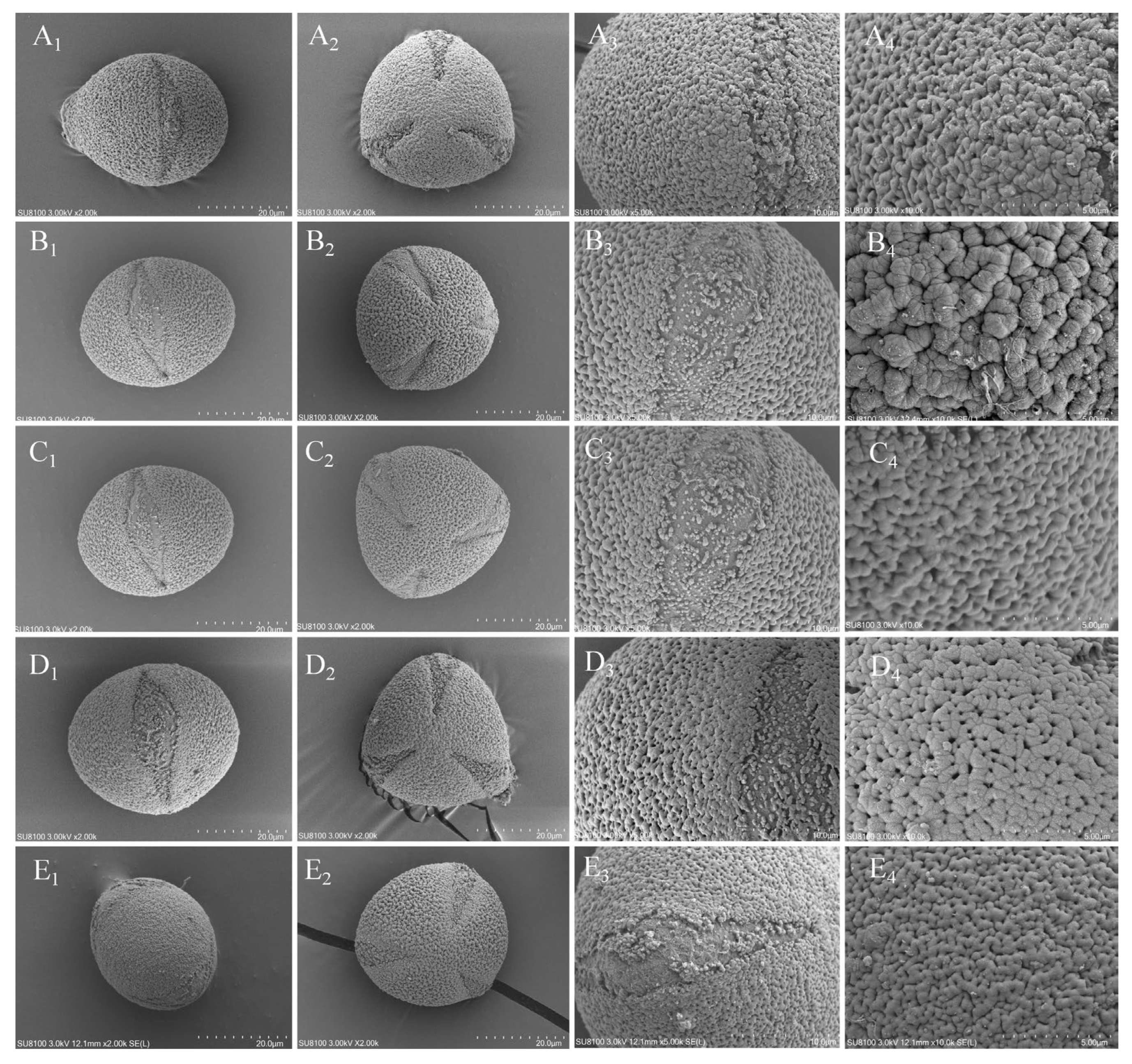
3.3. Features of Chloroplast Genomes
| acutiperulata | C. pyxidiacea | C. anlungensis | C. rubituberculata | C. leyeensis | |
|---|---|---|---|---|---|
| Genome size (bp) | 156,624 | 156,677 | 156,587 | 157,044 | 157,063 |
| GC (%) | 37.33 | 37.33 | 37.33 | 37.31 | 37.30 |
| LSC size (bp) | 86,212 | 86,261 | 86,262 | 86,689 | 86,661 |
| SSC size (bp) | 18,286 | 18,283 | 18,281 | 18,279 | 18,276 |
| IR size (bp) | 52,130 | 52,130 | 51,986 | 52,076 | 52,118 |
| GC in LSC (%) | 35.36 | 35.35 | 35.31 | 35.31 | 35.31 |
| GC in SSC (%) | 30.59 | 30.62 | 30.60 | 30.62 | 30.60 |
| GC in IR (%) | 42.95 | 42.95 | 42.96 | 42.98 | 42.96 |
| GC in CDS (%) | 37.61 | 37.53 | 37.54 | 37.65 | 37.55 |
| 1st position GC (%) | 45.37 | 45.19 | 45.24 | 45.42 | 45.26 |
| 2nd position GC (%) | 38.04 | 37.94 | 37.97 | 38.00 | 37.97 |
| 3rd position GC (%) | 29.43 | 29.43 | 29.40 | 29.53 | 29.40 |
| Length of CDS | 79,500 | 79,767 | 79,671 | 80,155 | 80,175 |
| Number of genes | 130 | 132 | 134 | 136 | 136 |
| Number of CDS | 87 | 87 | 89 | 91 | 91 |
| Number of tRNAs | 35 | 37 | 37 | 37 | 37 |
| Number of rRNAs | 8 | 8 | 8 | 8 | 8 |
| GenBank ID | OQ556869 | OP058659 | NC_050354 | MZ766253 | OK046127 |
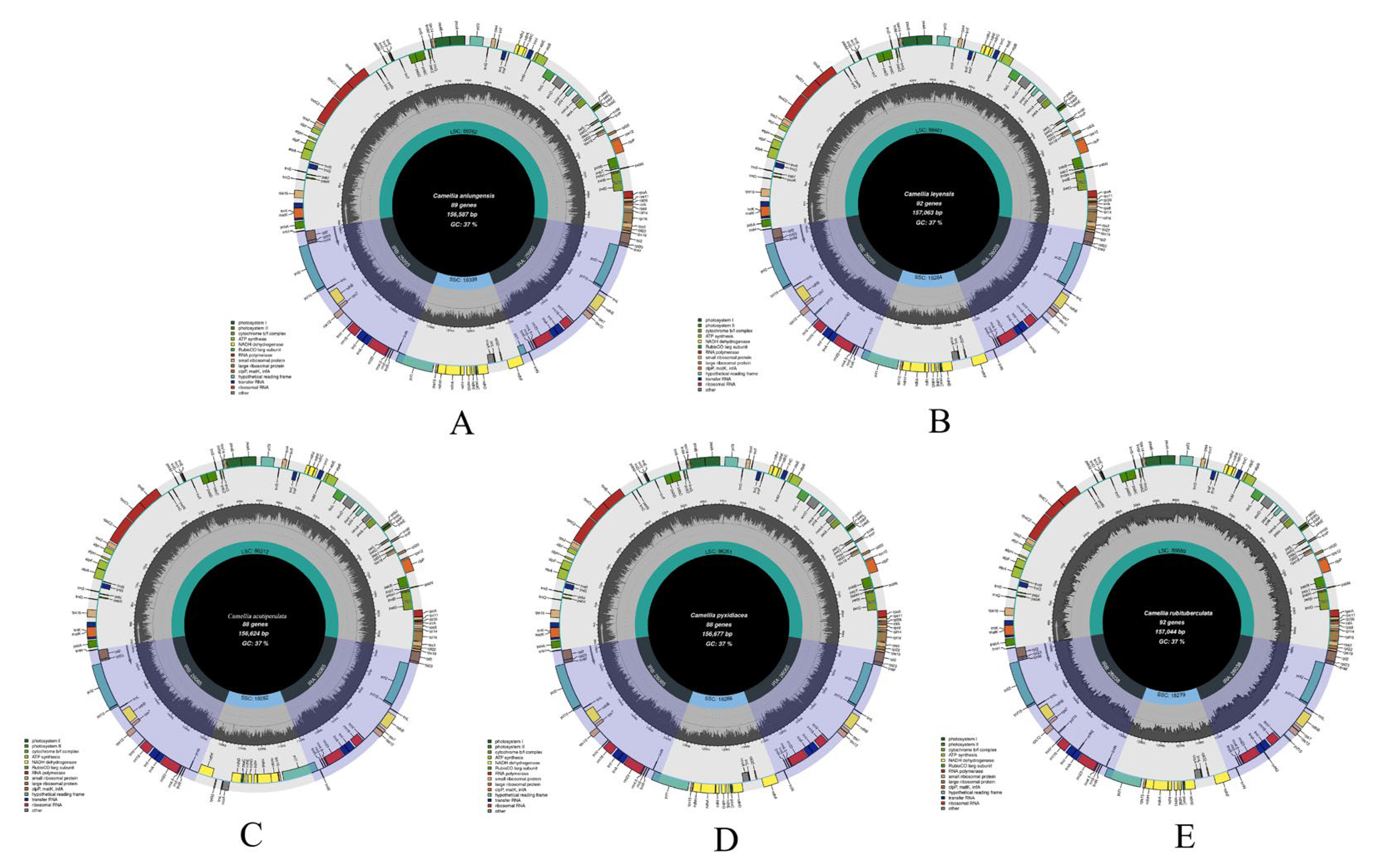
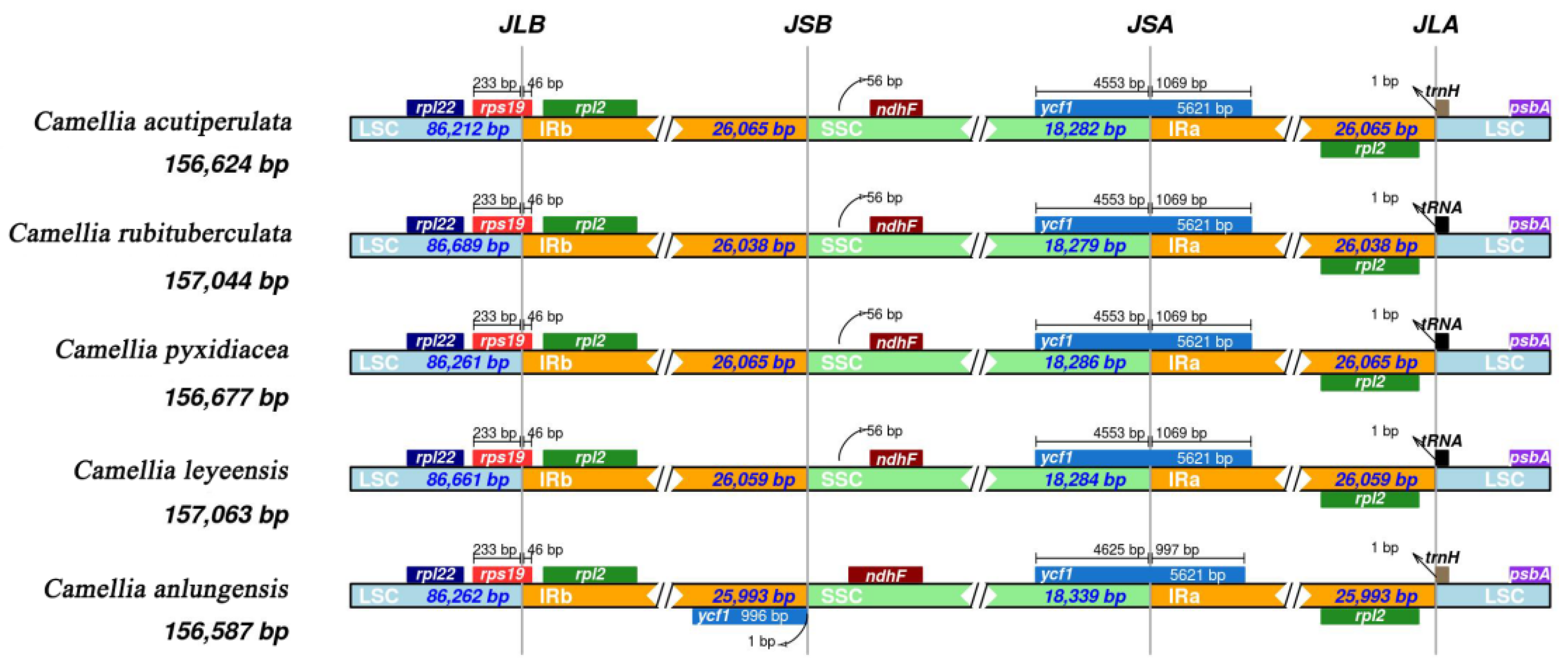
3.4. Phylogenetic Analysis
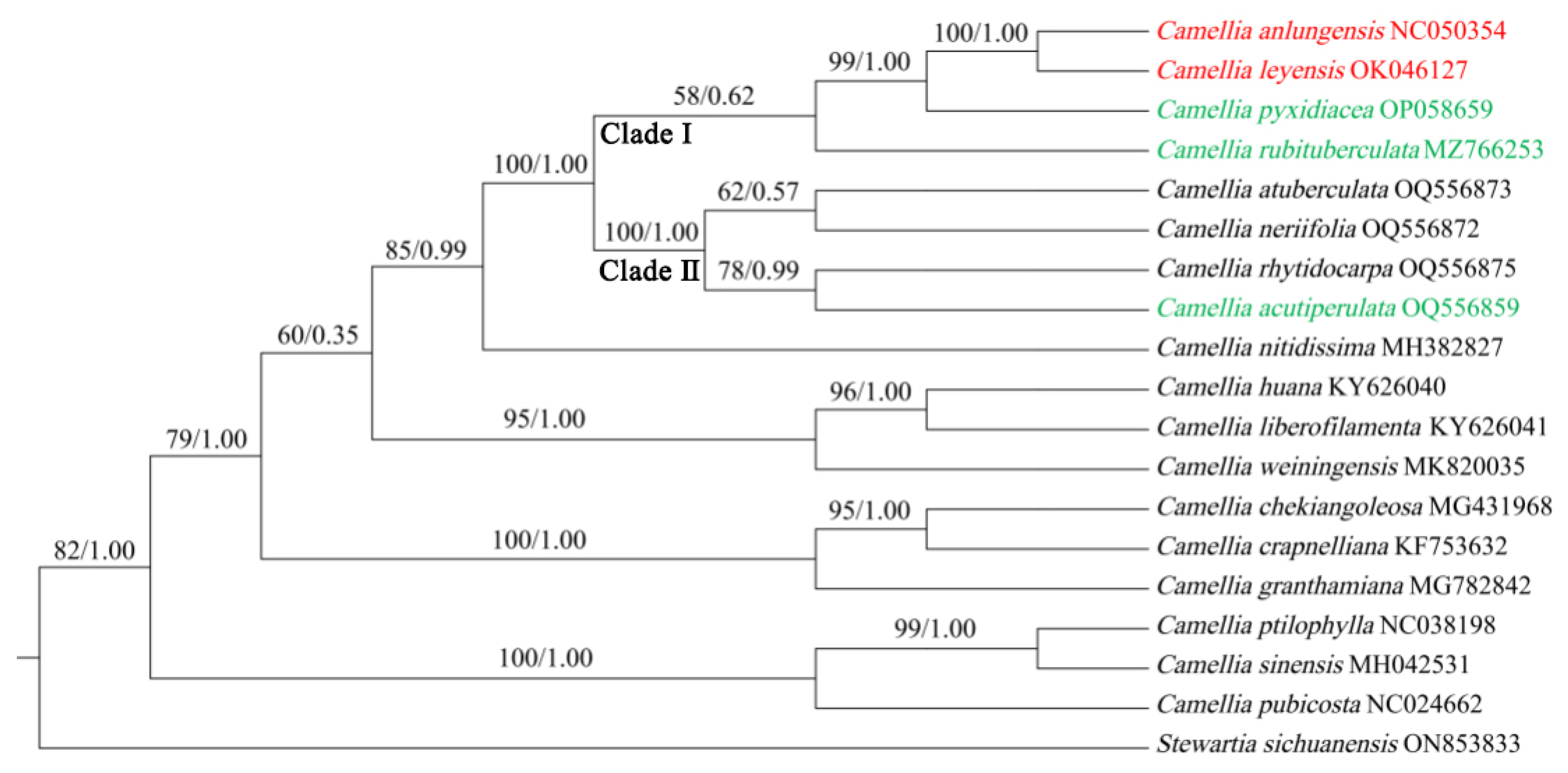
3.5. Taxonomic Treatment

Camellia Anlungensis Chang
Discussion
4.1. Taxonomic processing
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chang, H. T. A taxonomy of the genus Camellia. Editorial department of journal of Sun Yatsen University. 1981, 47–52. [Google Scholar]
- Chien, S.S. Four new ligneous plants of Szechuan. Contrib. Biol. Lab. Sci. Soc. China Bot. 1939, 12, 89–100. [Google Scholar]
- Min, T.L.; Zhong, Y.C. A revision of genus Camellia sect. Tuberculata. Acta Botanica Yunnanica. 1993, 15, 123–130. [Google Scholar]
- Min, T.L. A systematic synopsis of the genus Camellia. Acta Botanica Yunnanica. 1999, 21, 149–159. [Google Scholar]
- Chang, H.T.; Ren, S.X. Diagnosis on the systematic development of Camellia Ⅵ. revised on sect. Tuberculata of Camellia. supplement to the journal of Sun Yatsen University. 1996, 2: 55-60.
- Chang, H.T. New record of Camellia from South China. Acta Scientiarum Naturaliun Universitatis Sun Yatseni. 1984, 23, 77–82. [Google Scholar]
- Sealy. A revision of the genus Camellia. London: the Royal Horticulture Society. 1958, 1-239.
- Jiang, B.; Peng, Q.F.; Shen, Z.G.; Moller, M.; Pi, E.X.; Lu, H.F. Taxonomic treatments of Camellia (Theaceae) species with secretory structures based on integrated leaf characters. Plant Syst Evol. 2010, 290, 1–20. [Google Scholar] [CrossRef]
- Jiang, Z. D. Preliminary study of molecular phylogenetics and biogeography of the genus Camellia L. based on chloroplast DNA. Zhejiang Sci-Tech University. 2017.
- Wu, Q.; Tong, W.; Zhao, H.J.; Ge, R.H.; Li, R.P.; Huang, J.; Li, F.D. Comparative transcriptomic analysis unveils the deep phylogeny and secondary metabolite evolution of 116 Camellia plants. The Plant Journal. 2022, 111, 406–421. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X. Revision of the classification and comprehensive evaluation of the ornamental value of plants in the Camellia L. sect. Tuberculate. Guizhou University. 2023.
- Ran, Z.H.; Li, Z.; Xiao, X.; An, M.T.; Yan, C. Complete chloroplast genomes of 13 species of sect. Tuberculata Chang (Camellia L.): genomic features, comparative analysis, and phylogenetic relationships. BMC Genomics. 2024, 25, 108. [Google Scholar] [CrossRef]
- Lin, M.J.; Lu, Q.M. New record of Guizhou Camellia. Act. Sci. Nat. Univ. Sunyats. 1984, 83–85. [Google Scholar]
- Xu, Z.R.; Chen, F.; Deng, C.Y. A new species of sect. Tuberculata. Guihaia. 1987, 19–21. [Google Scholar]
- Chang, H.T.; Ren, S.X. A classification on the section Tuberculata of Camellia. Act. Sci. Nat. Univ. Sunyats. 1991, 30, 86–91. [Google Scholar]
- Erdtman, G. Handbook of palynology. Munksgaard, Copenhagen. 1978.
- Wang, K.F. & Wang X.Z. Introduction to sporology. Beijing: Peking University Press. 1983.
- Halbritter, D.D., Ulrich, D.S., Grímsson, D.F., Weber, P.D., Zetter, P.D., Hesse, P.D., Buchner, D.R., Svojtka, M.M., & Frosch-Radivo, A. Illustrated Pollen Terminology. Cambridge International Law Journal. 2018.
- Li, J.L.; Wang, S.; Yu, J.; Wang, L.; Zhou, S.L. A modified CTAB protocol for plant DNA extraction. Chinese Bulletin of Botany. 2013, 48, 72–78. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y. , Li, D.Z. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. Organellar Genome DRAW (OGDRAW): a tool for the easy generation of highquality custom graphical maps of plastid and mitochondrial genome. Current Genetics 2007, 52, 267–274. [Google Scholar] [CrossRef]
- Amyiryousefi, A.; Hyvonen, J.; Poczai, P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT: iterative refinement and additional methods. Methods Mol Biol. 2014, 1079, 131–46. [Google Scholar]
- Nguyen, L.T.; Schmidt, H.A.; Haeseler, A.; Minh, B.Q. IQTREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754-755. [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Research. 2021, 49, 293–296. [Google Scholar] [CrossRef]
- Efroni, I.; Eshed, Y.; Lifschitz, E. Morphogenesis of simple and compound leaves: a critical review. Plant Cell. 2010, 22, 1019–1032. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.X.; Zavada, M.S.; Min, T.L. Pollen morphology of Camellia (Theaceae) and ITS taxonomic significance. Acta Botanica Yunnanica. 1992, 14, 275–282. [Google Scholar]
- Hu, Z.M.; Zhao, C.H.; Zhao, Y.Y.; Liu, J.X. Pollen morphology of Liliaceae and its systematic significance. Palynology 2021, 45, 531–568. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




