1. Introduction
In recent years, global life expectancy has continued to rise, accompanied by a steady increase in the proportion of older adults in the population [
1,
2]. Aging is the most significant risk factor for numerous diseases, including cardiovascular disease, diabetes, neurodegenerative disorders, and cancer [
3,
4], and developing preventive strategies that target the aging process is considered a pressing challenge in both medical and nutritional sciences.
Various food-derived compounds with anti-aging potential have been identified [
5,
6]. Moreover, endogenous polyamines, such as putrescine (PUT), spermidine (SPD), and spermine (SPM), have attracted considerable attention because of their ability to promote cellular homeostasis and longevity [
7,
8]. SPD has demonstrated biological activities, such as anti-inflammatory effects [
9], autophagy induction, and deceleration of age-related cellular decline [
10]. These effects contribute to lifespan extension across diverse model organisms.
The naturally occurring polyamine content in conventional food sources varies widely but remains relatively low [
11,
12,
13,
14], making it difficult to intake sufficient polyamine solely through the diet. For instance, natto (fermented soybeans) contains approximately 190–680 nmol/g SPD [
12]. The average daily intake in the Japanese population is estimated to be 90 μmol PUT, 74 μmol SPD, and 36 μmol mg SPM per day [
14]. Polyamine levels tend to decline with age [
10], suggesting the need for exogenous supplementation to sustain cellular function and health. Thus, there is a clear demand for high-polyamine ingredients that can be used as functional food supplements. Therefore, we applied selective breeding techniques to
Saccharomyces cerevisiae sake yeast strains carrying polyamine biosynthetic genes to generate strains rich in SPD and SPM.
The core pathways of polyamine biosynthesis in yeast, such as those involving ornithine decarboxylase- and S-adenosylmethionine (SAM)-derived aminopropylation reactions, are well-characterized [
15,
16]. However, the specific genetic mutations and regulatory mechanisms that drive enhanced polyamine accumulation remain largely unexplored. Additionally, given the growing public concern over genetically modified organisms (GMOs) [
17], particularly among health-conscious consumers favoring clean-label products, our strain development strategy was deliberately based on traditional, non-GMO (non-GMO) methods. This classical breeding approach not only aligns with current consumer and regulatory preferences but also enhances the marketability and social acceptance of the resulting high-SPD yeast strain as a functional food ingredient.
Saccharomyces cerevisiae K7, originally isolated from sake brewing, was selected as the parental strain because of its robust fermentation performance and a long history of use in food production [
18].
Among the recognized anti-aging strategies, caloric restriction consistently extends lifespan across species, largely through enhanced autophagic activity [
19,
20]. Autophagy maintains cellular integrity by degrading and recycling damaged proteins and organelles, particularly under nutrient deprivation or stress [
21]. Beyond cellular housekeeping, it contributes to infection resistance, metabolic regulation, and longevity. In the skin, autophagy supports fibroblast renewal, extracellular matrix maintenance, and stress resilience, thereby promoting a healthy appearance and delaying aging [
22,
23]. Nutrients that activate autophagy are therefore of particular interest as dietary strategies to counteract age-related decline. Among them, autophagy-inducing compounds such as SPD have been linked to multiple health benefits. In preclinical models, autophagy enhancement delays neurodegeneration by improving proteostasis and reducing toxic aggregates, protects cardiovascular function through removal of damaged mitochondria, and preserves skeletal muscle integrity by facilitating cytoplasmic turnover [
24,
25].
In this study, we aimed to develop a non-GMO S. cerevisiae strain with enhanced SPD production and to evaluate its potential as a functional ingredient for health-promoting applications. We hypothesized that adaptive strain development would yield a yeast variant with increased polyamine accumulation and beneficial biological activities, including support for cellular proliferation and autophagy. To test this, we established a scalable fermentation system and conducted a comprehensive evaluation of the strain’s biochemical and functional characteristics. This work was designed to address the growing demand for safe, food-derived autophagy-inducing nutrients and to explore the potential of polyamine-enriched yeast as a functional ingredient. The findings provide a foundation for the application of SPD-rich yeast in promoting skin health and broader human health benefits.
2. Materials and Methods
Nutritional composition analysis (
Table S1), Safety and quality evaluation (
Table S2,
Table S3), whole-genome sequencing and free amino acid profiling were outsourced to certified external laboratories, and analytical procedures were conducted in accordance with each laboratory's internal quality standards. The methods are detailed in the Supplementary Information.
2.1. Yeast Strain Development
The parent yeast strain (S. cerevisiae K7) was cultured in liquid medium containing 1% Bacto yeast extract, 2% Bacto peptone, and 2% glucose at 30 °C for 16 h with 210 rpm shaking. The culture was exposed to 45 W ultraviolet light at a dose sufficient to achieve ~1% survival, and then diluted 104–105-fold with 50 mM potassium phosphate buffer (pH 7). Treated yeasts were spread onto agar plates containing the same nutrient composition and 2% agarose and incubated at 30 °C for 48 h. Colonies were screened for high polyamine content using HPLC. One mutant strain, designated as 3L63, demonstrated a significant increase in SPD content and was selected for further characterization and scaling up.
2.2. Yeast Fermentation and Drying
A high-polyamine-producing mutant yeast (3L63 strain) was cultured in a medium (Japanese Patent 6663598). The yeast was cultured at 30 °C for 4 d with agitation. After cultivation, the broth was centrifuged at 8000rpm, 4 °C for 10min to concentrate the yeast cells, which were then washed with water. The resulting mixture was then incubated at 60 °C for 30 min. The yeast was then spray-dried using a Büchi Mini Spray Dryer B-290 (BÜCHI Labortechnik AG, Flawil, Switzerland) at an outlet temperature of 95–100 °C to obtain dried yeast powder. The culture conditions were scaled up into a 3-L jar and then a 30-L jar with the same medium composition and at the same temperature (30 °C). The aeration and agitation conditions were adjusted according to the vessel scale.
2.3. Polyamine Quantification Through HPLC
Perchloric acid (60%), sodium carbonate, acetone, L-proline, toluene, and acetonitrile were purchased from Wako Pure Chemical Industries (Osaka, Japan). Dansyl chloride (10%) in acetone was purchased from the Tokyo Chemical Industry (TCI) (Tokyo, Japan). SPM phosphate hexahydrate, 1,7-diaminoheptane (TCI), SPD trihydrochloride (Sigma-Aldrich, St. Louis, MO, USA), and PUT dihydrochloride (Nacalai Tesque, Kyoto, Japan) were used as standards for SPM, SPD, and PUT, respectively. They were prepared in the same extraction solvents and processed as the samples to generate calibration curves.
The workflow for sample preparation and polyamine derivatization is shown in
Figure S1. Powdered yeast samples (10–15 mg) were weighed into 15-mL tubes and mixed with 4 mL of extraction solvent containing 2 ppm 1,7-diaminoheptane (internal standard) in 5% perchloric acid. The samples were vortexed and incubated at 50 °C with shaking at 125 rpm for 30 min. After extraction, 1 mL of the supernatant was transferred to 1.5-mL microtubes and centrifuged at 12,000 rpm for 5 min. A 100-µL aliquot of the clear supernatant was diluted 5-fold with 400 µL of extraction solvent and placed into glass centrifuge tubes.
Subsequently, 250 µL of 18% sodium carbonate solution and 500 µL of 10 g/L dansyl chloride in acetone were sequentially added. The mixture was allowed to react at 45 °C for 1 h under light-shielded conditions. After cooling, 250 µL of 10% proline was added to quench the excess dansyl chloride, followed by a 10 min reaction. Subsequently, 2 mL of toluene was added for phase extraction. After mixing and centrifugation (2,000 rpm, 5 min, 25 °C), 1 mL of the organic layer was collected and dried under a nitrogen atmosphere at room temperature (25 °C). The residue was re-dissolved in acetonitrile (0.5 mL) via ultrasonication for 40 min and then used for HPLC analysis.
Polyamines were separated through a C18 column (YMC-Pack ODS-A, 4.6 × 150 mm, 5 μm; YMC, Kyoto, Japan) and analyzed using an HPLC system with Shimadzu LabSolutions software Shimadzu Corp., Kyoto, Japan). The mobile phase consisted of acetonitrile and water (72:28, v/v) at a flow rate of 1.5 mL/min. The column was maintained at 40 °C, and the injection volume was 20 µL. Fluorescence was detected at excitation and emission wavelengths of 365 and 510 nm, respectively. The total run time was 30 min per sample.
2.4. Whole-Genome Sequencing, Mutation Detection, and Functional Enrichment Analysis
Genomic DNA was extracted from the selected 3L63 yeast strain using the Dr. GenTLE (from Yeast) High Recovery kit (Takara Bio, Shiga, Japan), according to the manufacturer’s protocol. Whole-genome sequencing and mutation analyses were outsourced to Hokkaido System Science Co., Ltd. (Sapporo, Japan), as described in the Supplementary Information. Genes with high and moderate annotations were identified from the results of the gene mutation analysis. Next, Gene Ontology (GO) and pathway enrichment analyses of the listed genes were performed using g:Profiler [
26] (
https://biit.cs.ut.ee/gprofiler) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Mapper [
27] (
https://www.genome.jp/kegg/mapper/) web tools, providing insight into their roles in metabolic networks and potential links to polyamine biosynthesis.
2.5. Free Amino Acid Analysis
For amino acid analysis, K7 and 3L63 yeast strains were cultured in YEPD medium composed of 10 g/L yeast extract, 20 g/L Bacto peptone, and 20 g/L glucose. Pre-cultures were prepared by inoculating each strain into 4 mL of YEPD medium in test tubes and incubating overnight at 30 °C. For the main culture, 500 mL of YEPD medium was dispensed into sterile 1-L Erlenmeyer flasks. Using an aseptic technique in a biosafety cabinet, pre-culture of each strain from one test tube was inoculated into triplicate flasks containing 500 mL of culture media. The cultures were incubated at 30 °C with shaking at 100 rpm for 20 h. After incubation, the yeast cultures were transferred into 500-mL centrifuge bottles and centrifuged at 8000 rpm for 10 min to pellet the cells. The supernatants were discarded, and the pellets from each strain were pooled and resuspended in 250 mL 0.9% sodium chloride solution. After centrifugation at 500×g, 25 °C for 15min, the supernatant was discarded. Ethanol (200 mL) was added to the pellet to wash the cells, followed by centrifugation and removal of the supernatant. The final yeast pellets were dried under reduced pressure at 50 °C overnight and then air-dried at 25 °C for 3 d to obtain dried yeast samples for analysis.
Amino acid analysis was outsourced to IDEA Consultants, Inc. (Osaka, Japan), and the free amino acids in the K7, 3L63, and 3L63 yeast products were analyzed using HPLC. The sample preparation and analysis procedures are described in the Supplementary Information.
2.6. HDF Culture and Cell Proliferation Assay
We evaluated the effect of 3L63 yeast on the proliferation of HDFs provided by Kurabo Industries, Ltd. (Osaka, Japan). The cells were maintained in a culture medium (Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin). For the proliferation assay, HDFs were seeded in 96-well plates and treated with different concentrations of 3L63 yeast in the control media. In a subset of experiments, to mimic aging conditions, the cells were treated with 400 μM hydrogen peroxide for 2 h (H2O2; Wako Pure Chemical Industries) to induce senescence by oxidative stress before adding 3L63 yeast at several concentrations. After 24 h of incubation, cell viability and proliferation were assessed using Cell Counting Kit-8 (Dojindo, Tokyo, Japan). The absorbance was measured at 450 nm using a microplate reader (Multimode Plate Reader ARVO X3; PerkinElmer, Shelton, CT, USA).
2.7. Autophagy Induction Analysis
2.7.1. CYTO-ID Staining for Autophagic Activity
HeLa cells were cultured in DMEM supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 µg/mL streptomycin. For the CYTO-ID autophagy detection assay (Enzo Life Sciences, Inc., New York, USA), cells were seeded at a density of 3 × 10
4 cells/well in a 24-well plate and incubated at 37 °C with 5% CO
2 for 24 h. On the following day, the culture medium was removed and replaced with 0.75 mL of fresh medium. Then, 0.25 mL of the test sample, prepared at four times the final concentration, was added to each well, resulting in a total volume of 1 mL. After incubation for 4.5 h at 37 °C and 5% CO
2, the cells were washed twice with 2 mL of 1× assay buffer. Subsequently, 0.5 mL of staining solution, composed of 2 µL CYTO-ID Green Detection Reagent 2 and 1 µL Hoechst 33342 Nuclear Stain diluted in 1 mL of culture medium, was added to each well. The cells were incubated in the dark at 37 °C for 30 min. After staining, the cells were washed once with 1 mL of 1× assay buffer and once with 1 mL culture medium. Finally, the medium was replaced with 0.5 mL of PBS, and the cells were imaged under a fluorescence microscope (Nikon Eclipse TC2000-S; Nikon, Japan) equipped with a digital camera (Nikon Ds-Ri2). The intensity of CYTO-ID fluorescence and number of nuclei were quantified using ImageJ software [
28].
2.7.2. mRFP-GFP Tandem Fluorescent-Tagged LC3 (tfLC3) Assay
Yeast samples were treated with artificial digestion fluid (Disintegration Test Solution No.1 containing 0.2% pepsin; Wako Pure Chemical Industries and Disintegration Test Solution No.2 containing 0.28% pancreatin; Wako Pure Chemical Industries). An aqueous extract was prepared by soaking the samples in distilled water. The tfLC3 assay was performed by Human Metabolome Technologies, Inc. (Yamagata, Japan), following the method described in [
29] and the Supplementary Information.
2.8. Protein Variant Modelling and Docking Simulation
The variant protein structure was predicted using protein structure modelling (SWISS-MODEL) via a homology modeling approached [
30]. The parent strain K7 and variant strain 3L63 protein sequences were used as the input for structure prediction. The quality of the predicted models was evaluated based on GMQE and QMEAN scores provided by SWISS-MODEL.
Docking simulations were performed using AutoDock Vina through the CB-Dock2 platform [
31]. This method automatically detects potentials binding cavities on the protein surface and carried out docking within the identified sites. For each simulation, CB-Dock2 provided the cavity volumes and the corresponding Vina score, which were used to evaluate the binding affinity and binding pocket characteristics. The best-ranked docking poses were selected on the lowest Vinal score and biologically relevant binding volume. These results were used to compare between the same proteins derived from the parent and variant strains. This comparison allowed assessment of how the introduced mutation altered ligand-binding properties and potential functional differences between the two protein forms.
2.9. Statistical Analysis
Statistical analyses were performed using SigmaPlot 15.0 or Excel software. One-way analysis of variance (ANOVA) was used for all pairwise multiple comparisons using Tukey’s honestly significant difference (HSD) test or the paired t-test for appropriate experiments.
3. Results
3.1. Physical Characterization of Newly Developed 3L63 Yeast
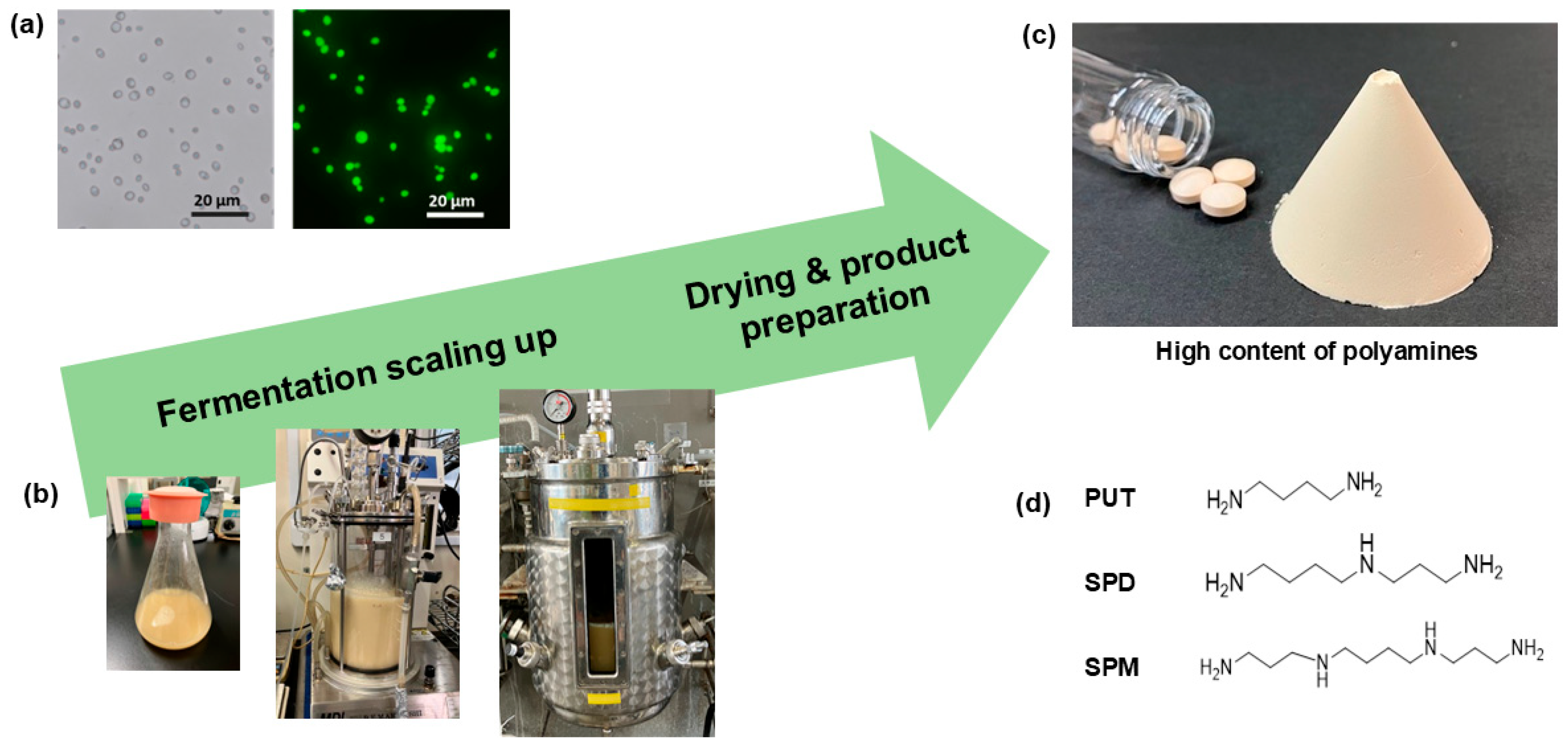
A high–polyamine-producing
S. cerevisiae strain was generated through ultraviolet mutagenesis of the K7 strain. The selected mutant, designated 3L63, was scaled up to the commercial production level (
Figure 1). The strain was successfully cultivated in a 30-L jar fermenter, and the resulting cells were washed, sterilized, and spray-dried to obtain an SPD-enriched yeast powder.
Figure 1 shows the morphology of the yeast under a microscope and the appearance of the final powdered product. The dried yeast was off-white to light-yellow in color and insoluble in water.
The nutritional profiles of the 3L63 yeast-derived powders are summarized in
Table 1. The parent
S. cerevisiae K7 strain is food-safe, and the 3L63-derived powder passed acute oral toxicity and hygiene tests in accordance with the Japanese food safety standards (
Table S2). Comprehensive quality assessments confirmed that the powder contains ≥2.0 mg/g of SPD, with a moisture content ≤6.0%, and no detectable foreign matter. Microbiological analysis showed total viable bacteria ≤300 CFU/g and fungi ≤100 CFU/g; coliforms were not detected. Heavy metals (Pb) and arsenic (As
2O
3) were within acceptable limits (≤20 ppm and ≤1 ppm, respectively). These results support the chemical and microbiological safety of these products in potential food and nutraceutical applications. The product specifications listed in
Table S3 were established to ensure product safety and consistency. For the sub-end-product, the yeast was mixed with 15% α-cyclodextrin post-process, as described in the Supplementary Information.
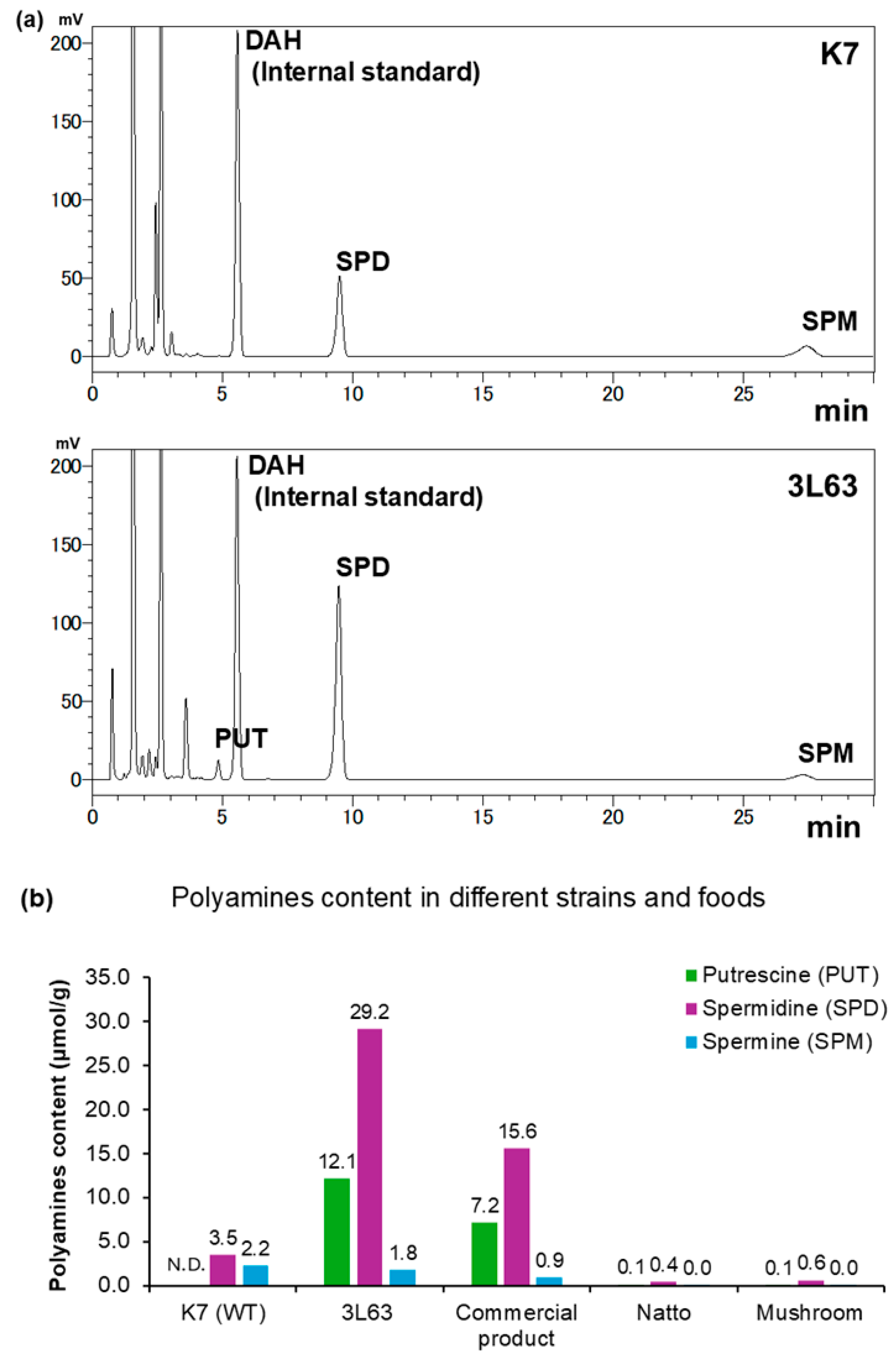
3.2. Polyamine Content in Yeast Strains
To evaluate the polyamine-producing capacity of the newly developed yeast strain 3L63, we quantified the intracellular levels of three major polyamines: PUT, SPD, and SPM. The polyamine content of 3L63 was compared with that of the parent K7 strain and its commercial yeast product. To obtain a commercial product, the formulation was optimized for improved powder handling and consistent production, achieved through modifications to the culture conditions and addition of cyclodextrin. Although the underlying yeast strains were identical, these alterations resulted in distinct polyamine profiles.
The parent K7 strain exhibited low levels of SPD (0.51 mg/g) and SPM (0.45 mg/g), with PUT below the detection limit, resulting in a total polyamine content of only 0.96 mg/g (
Table 2 and
Figure 2a). In contrast, the 3L63 strain accumulated significantly higher levels of polyamines, with 1.07 mg/g PUT, 4.24 mg/g SPD, and 0.36 mg/g SPM, amounting to 5.66 mg/g. This reflects an approximately 5.9-fold increase compared with that of the parent strain.
The commercial yeast product contained 0.63 mg/g PUT, 2.26 mg/g SPD, and 0.19 mg/g SPM, with total polyamine content of 3.08 mg/g. The final product maintained a considerably higher polyamine content than K7 yeast. Additionally, we compared polyamine levels in 3L63 cells with those reported in natural food sources, such as natto (fermented soybeans) [
12] and mushrooms [
13] (
Figure 2b). These findings demonstrate that 3L63 is a robust, high-polyamine-producing strain that is especially rich in SPD and is suitable for application as a functional ingredient in foods and supplements.
3.3. Whole-Genome Sequencing and Functional Enrichment Analysis
Whole-genome sequencing of S. cerevisiae 3L63 yielded 43,539,176 reads, of which 80.59% (35,089,430 reads) were successfully mapped to the reference genome of the K7 strain. Approximately 2.80% (1,220,805 reads) of the mapped reads were identified as duplicates, whereas 19.41% (8,449,746 reads) remained unmapped. After variant calling and annotation, genetic variants were classified based on their predicted functional impact. Only variants with high and moderate effects were selected for downstream analysis, including missense mutations, nonsense mutations, and frameshift variants, which are likely to alter protein function and are thus considered biologically meaningful.
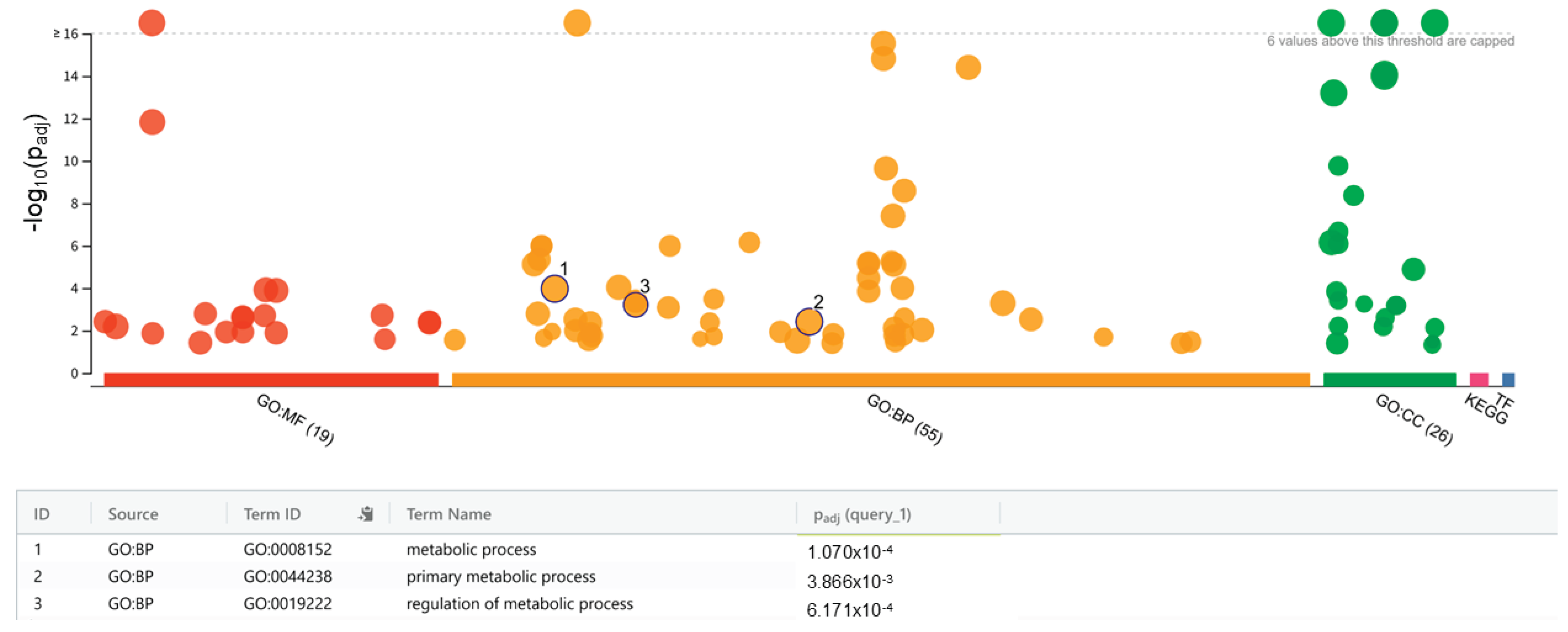
To evaluate the biological implications of the identified variants, functional enrichment analysis was performed using a filtered gene list (860 genes) and the g:Profiler tool. A total of 55 significantly enriched biological process (BP) terms, 19 molecular function (MF) terms, and 26 cellular component (CC) terms were identified (adjusted
p < 0.05) (
Figure 3). The significant BP terms also included the regulation of metabolic processes (GO:0019222, adjusted
p = 6.171 × 10
-4), primary metabolic processes (GO:0044238, adjusted
p = 6.171 × 10
-4), and metabolic processes (GO:0008152, adjusted
p = 1.070 × 10
-3). This result suggests that the variants are involved in the regulation of core metabolic pathways and that these mutations may alter the ability of the yeast strain to synthesize, degrade, or regulate key metabolites. The detailed results are presented in
Figure S2.
KEGG pathway mapping of the filtered gene list revealed hits for the pathways listed in
Table S4. These pathways suggest potential rewiring of central metabolism and amino acid biosynthesis, which could affect cellular growth and stress adaptation.
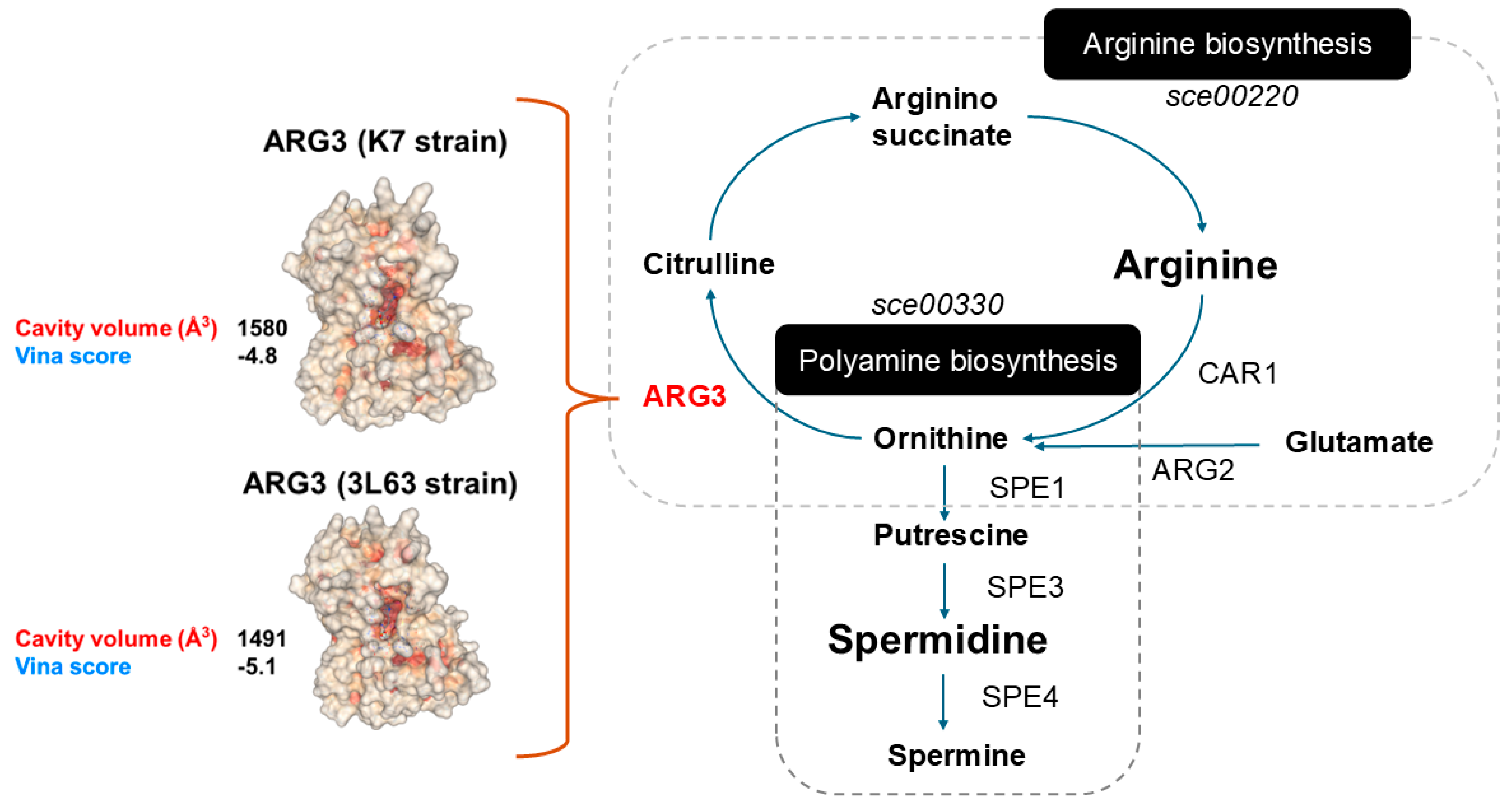
Manual inspection of the gene list revealed five mutations in central metabolic pathway genes:
SAM1, LEU2, ACO2, ARG3, and AGC1 (
Table S5). ARG3 is functionally associated with arginine metabolism, which occurs upstream of PUT and is a major polyamine. However, polyamine-specific GO terms (e.g.,
GO:0006596: polyamine biosynthetic process) were not significantly enriched, possibly because of the limited number of direct polyamine biosynthesis genes in the input list. To further investigate the potential impact of the ARG3 mutation, we compared the protein structures of ARG3 K7 and ARG3 3L63 using in silico modeling. The analysis suggested that the mutation may alter the cavity volume for the ARG3 substrate ornithine and potentially enhance substrate binding, as indicated by improved docking scores from AutoDock Vina (
Figure S3).
3.4. Free Amino Acid Analysis
Intracellular polyamine levels were quantified in K7 and 3L63 yeast strains to assess polyamine dynamics during cultivation (
Table 3). The total polyamine content in K7 was 2.07 mg/g, comprising 0.04 mg/g PUT, 1.62 mg/g SPD, and 0.41 mg/g SPM. Consistently, the 3L63 strain exhibited a markedly higher polyamine level of 4.17 mg/g, consisting of 0.39 mg/g PUT, 3.41 mg/g SPD, and 0.37 mg/g SPM. This approximately two-fold increase was primarily attributed to the enhanced SPD accumulation.
Intracellular free amino acid profiles were examined during yeast cultivation. As an increase in free amino acids can result from protein degradation, ethanol was applied immediately after cultivation to fix the cells and suppress autolysis. The amino acid content of the final product was assessed using samples subjected to different cultivation times and sterilization processes. The results are presented in
Table 3 and
Tables 3 and S4.
Overall, the total free amino acid content of the 3L63 strain (283 μmol/g) was slightly lower than that of the K7 parent strain (306 μmol/g). Despite this, several individual amino acids showed significant variation. Arginine nearly doubled in 3L63, increasing from 17.4 μmol/g (5.7%) to 28.3 μmol/g (10.0%), suggesting enhanced biosynthesis or accumulation in the new variant. Lysine also increased from 6.10 μmol/g (2.0%) in K7 to 12.9 μmol/g (4.6%) in 3L63. Conversely, glutamic acid decreased from 134 μmol/g (43.8%) in K7 to 104 μmol/g (36.7%) in 3L63, although it remained the most abundant amino acid in both strains. Minor differences were also observed in glycine (K7:10.4 μmol/g; 3L63:13.8 μmol/g), alanine (K7:14.9 μmol/g; 3L63:10.5 μmol/g), and serine (K7:43.8 μmol/g; 3L63:43.5 μmol/g), whereas the overall amino acid profile remained relatively consistent. Notably, the levels of branched-chain amino acids (leucine, isoleucine, and valine) were lower in 3L63 cells than in K7 cells.
A comparison between the mid-fermentation sample of 3L63 and the final 3L63-derived product revealed a substantial increase in total amino acids from 283 to 354 μmol/g. This suggests a concentration effect or selective enrichment during downstream processing. Arginine exhibited a considerable increase from 28.3 μmol/g (10.0%) to 65.1 μmol/g (18.4%), becoming a dominant amino acid in the final product. Alanine also increased sharply from 10.5 μmol/g (3.7%) to 38.9 μmol/g (11.0%). Significant increases were also observed in branched-chain amino acids, with valine increasing from 1.83 to 15.2 μmol/g, isoleucine from 1.17 to 13.6 μmol/g, and leucine from 1.90 to 19.4 μmol/g.
Glutamic acid decreased from 104 μmol/g (36.7%) to 43.9 μmol/g (12.4%), and serine from 43.5 μmol/g (15.4%) to 12.7 μmol/g (3.6%) during fermentation, indicating selective degradation or metabolic transformation. Additionally, the final product showed marked increases in aromatic and sulfur-containing amino acids, including phenylalanine (from 0.55 to 10.2 μmol/g), tyrosine (from 0.37 to 9.51 μmol/g), cystine (from 4.41 to 9.31 μmol/g), and methionine (from 2.79 to 6.10 μmol/g), thereby enhancing its nutritional value. Overall, transformation from the parent yeast K7 to the 3L63 strain to the final product resulted in pronounced enrichment of essential amino acids and a notable increase in polyamine content, particularly SPD, underscoring the potential of this strain for developing nutritionally enhanced yeast-based products.
3.5. HDF Proliferation
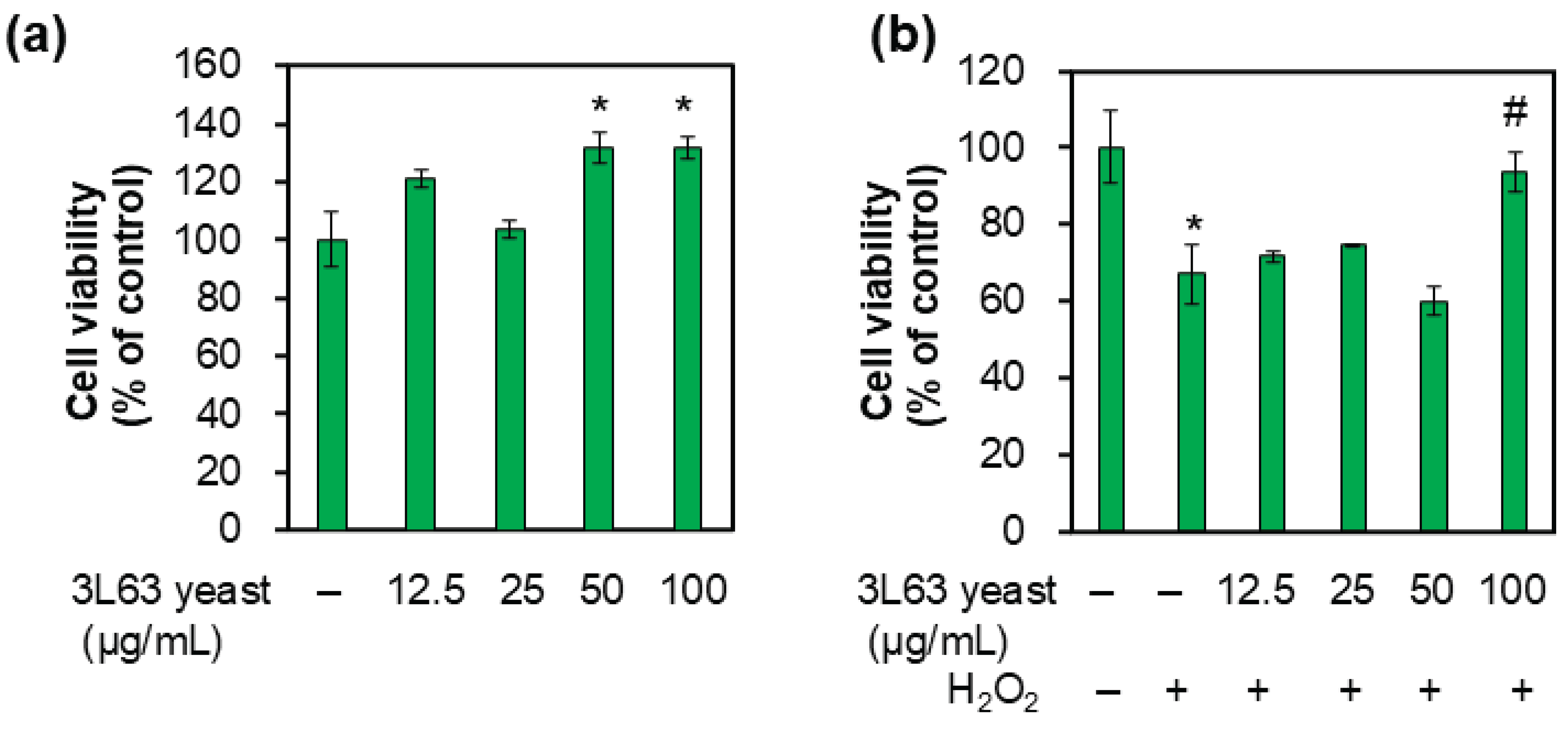
We evaluated whether the proliferative ability of fibroblasts, vital for skin health, can be enhanced by the addition of SPD-enriched yeast. Our experiments confirmed that 3L63 yeast significantly promoted cell proliferation not only in normal fibroblasts but also in senescence-induced cells with 100 μg/mL yeast, equivalent to 1.72 μM SPD supplementation (
Figure 4). These findings suggest that SPD-enriched 3L63 yeast may help restore the diminished proliferative capacity of aging fibroblasts and improve signs of skin aging.
3.6. Autophagy Induction by 3L63 Yeast
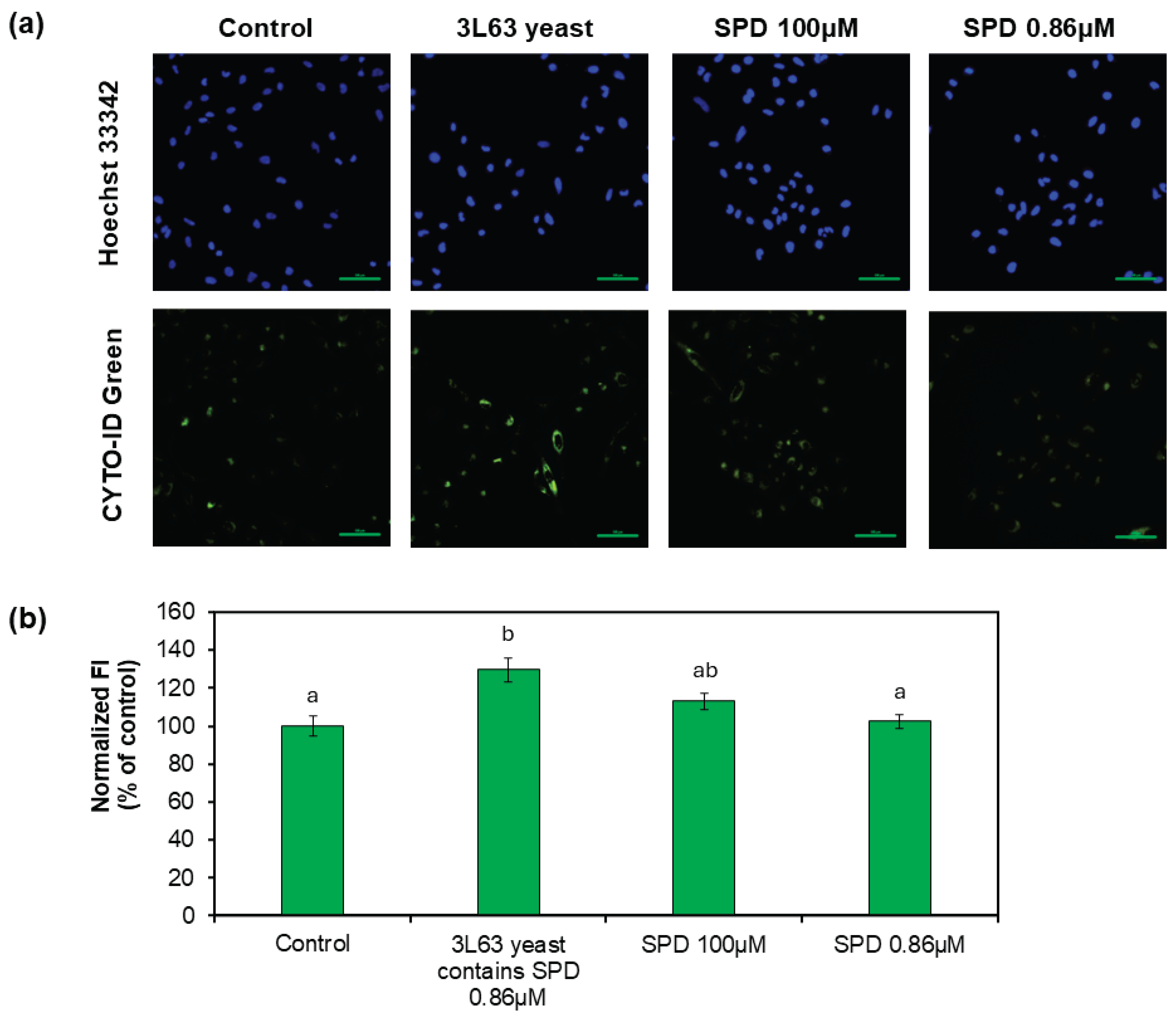
Autophagy activation was first assessed in HeLa cells using the CYTO-ID Autophagy Detection Kit (
Figure 5a, b). Fluorescence intensity analysis revealed that treatment with 3L63 yeast (containing 0.86 μM SPD) significantly enhanced autophagic activity, reaching an average fluorescence intensity of 130% relative to the untreated control (
p < 0.001). Notably, this effect was substantially greater than that observed with pure SPD treatment. Specifically, 100 μM pure SPD induced a modest increase to 113% of control levels, whereas 0.86 μM SPD showed only a slight elevation to 102%, neither of which were significant differences compared to the control group. These findings suggest a potential synergistic effect between SPD and other bioactive components present in the yeast matrix, which may stimulate autophagy more effectively than SPD alone.
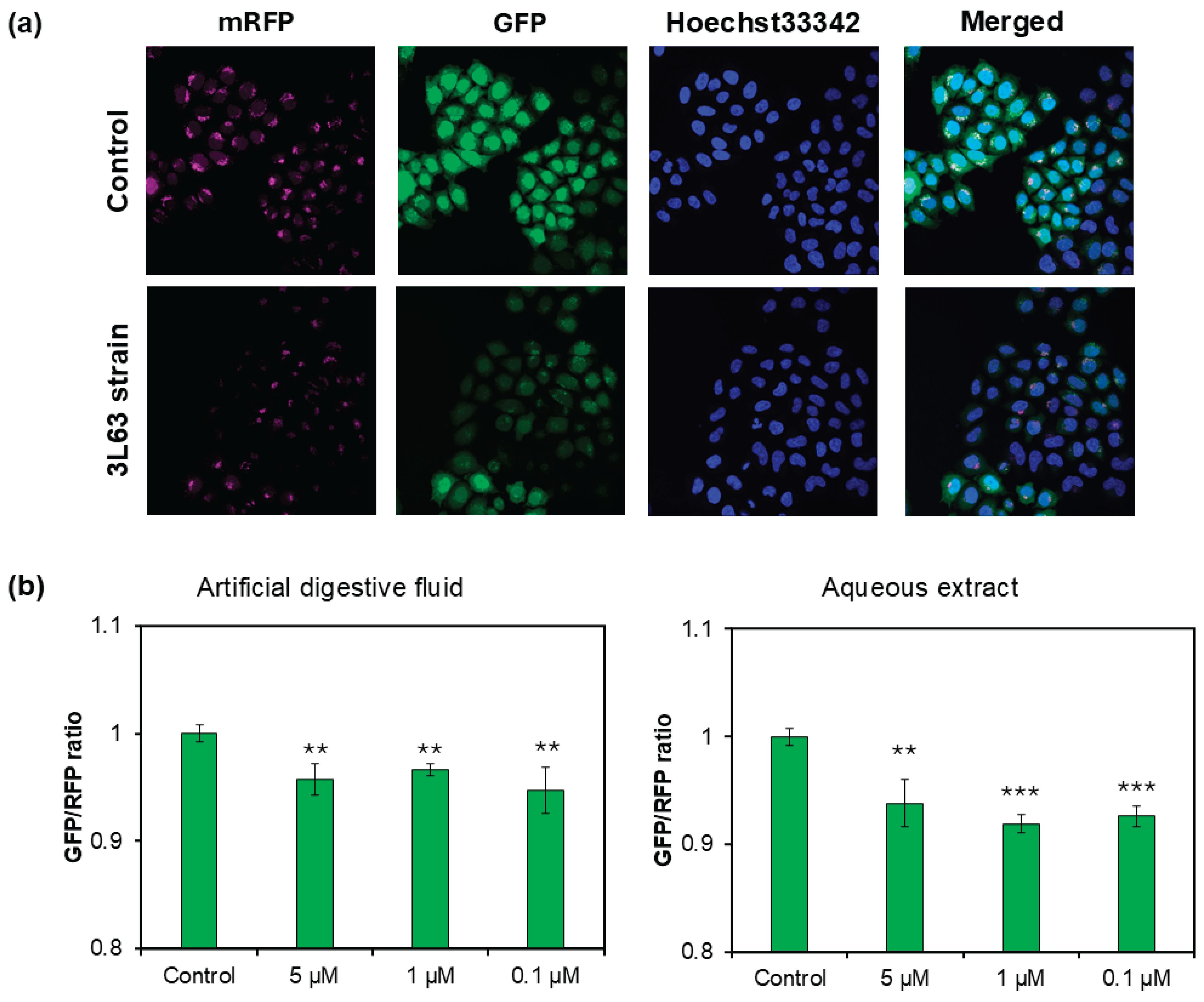
To validate our observations regarding autophagy induction, a tandem fluorescent-tagged LC3 (tfLC3) assay was performed using HeLa cells (
Figure 6a). The cells were treated with aqueous extracts and artificial digestion fluid-treated 3L63 yeast corresponding to 0.1, 1, and 5 μM SPD concentrations. Even at the lowest dose (0.1 μM SPD equivalent), a significant increase in autophagy was observed compared to that in the control, supporting the robust activity of the yeast product. Additionally, the levels of autophagy-related protein LC3-II were examined in HDFs using western blot analysis under autophagic flux conditions (
Figure S5). Although the result was based on a single experiment (no replicates were performed), the results consistently showed that 3L63 yeast containing 50 μM SPD induced higher LC3-II expression than pure SPD and starvation conditions, further supporting the superior autophagy-inducing capacity of the yeast-derived SPD formulation.
4. Discussion
4.1. Food Safety and Formulation Strategy
Yeast is considered GRAS (Generally Recognized as Safe) by regulatory authorities such as the U.S. FDA and has a long-established history of use in fermented foods, including bread, beer, and dietary supplements [
32]. The parent strain used in this study,
S. cerevisiae K7, was originally isolated from sake brewing and is well-known for its robust fermentation performance, high ethanol tolerance, and adaptability to various stress conditions. As a diploid homothallic strain, K7 has been extensively used in traditional Japanese food production. Genomic analyses have revealed that K7 harbors unique alleles related to flavor compound synthesis, nitrogen metabolism, and stress response, which contribute to its desirable brewing characteristics [
18,
33,
34]. Furthermore, transcriptomic and proteomic studies have provided insights into K7’s metabolic regulation under fermentation conditions, thereby highlighting its potential as a platform for the development of functional food ingredients [
35,
36].
Based on this robust and well-characterized background, we developed a food-grade non-GMO yeast strain (3L63) that is enriched in polyamines, especially SPD, using classical mutagenesis and selection. Previous studies have enhanced polyamine production in yeast primarily through genetic engineering strategies. For example, Kim et al. (2017) successfully engineered
S. cerevisiae to overexpress SPD biosynthetic enzymes, achieving notable increases in SPD production at laboratory scale [
37]. Similarly, Qin et al. (2021) established a comprehensive metabolic engineering platform to produce SPD and polyamine analogues at high titers, demonstrating the potential of engineered yeast for both natural and unnatural polyamine production [
38].
While these engineered strains achieved high-level production, they involve genetic modifications, which may pose regulatory and consumer acceptance challenges for food applications. In contrast, our classical mutagenesis approach generated a non-GMO strain that complies with food-grade regulations and maintains market acceptability. Compared to the engineered strains, 3L63 demonstrated stable SPD accumulation under diverse nutrient and oxygenation conditions and was successfully scaled up from shake flasks to 3-L and 30-L fermenters, and ultimately to industrial volumes exceeding 10,000 L. To obtain a product ready for commercial use, we supplemented the strain with cyclodextrin to make the powder easier to handle and enhance its functionality [
39,
40].
Nevertheless, limitations of the mutagenesis approach remain. Random mutations make it challenging to precisely identify the genetic changes responsible for the enhanced SPD phenotype. Additionally, long-term evolutionary stability during repeated industrial propagation requires further validation. Despite these considerations, 3L63 represents a practical and regulatory-compliant strategy for producing SPD-enriched yeast, complementing prior metabolic engineering efforts reported [
37,
38]
4.2. Mechanism of Enhanced SPD Production
To elucidate the mechanisms underlying this phenotype, we combined whole-genome sequencing, amino acid profiling, and metabolic pathway interpretation. Whole-genome variant analysis revealed several mutations in genes associated with amino acid metabolism and central metabolic regulation, including
ARG3,
SAM1,
LEU2,
ACO2, and
AGC1. Among these,
ARG3, which encodes an ornithine carbamoyltransferase, plays a pivotal role in the urea cycle and catalyzes the conversion of ornithine and carbamoyl phosphate to citrulline, an essential step in arginine biosynthesis [
41,
42]. A moderate-impact missense mutation was identified in
ARG3 of 3L63 strain. Structural modeling and molecular docking analysis revealed a reduction in the substrate-binding cavity volume (from 1580 to 1491 ų) and an improved Vina binding score (from –4.8 to –5.1), suggesting enhanced binding affinity of ornithine to the mutant enzyme. Although ornithine levels were not directly quantified in this study, this finding implies an improved conversion efficiency from ornithine to citrulline, supporting an increased arginine biosynthetic flux.
Free amino acid profiling further supported this hypothesis, showing significantly elevated levels of arginine, lysine, glycine, and aspartic acid in 3L63 cells, whereas methionine and glutamic acid levels remained unchanged or decreased. The accumulation of arginine may reflect altered regulation of nitrogen metabolism and urea cycle flux, potentially mediated by the
ARG3 mutation. This increase in the upstream precursor pools likely fuels the native polyamine biosynthetic pathway in yeast; arginine is converted to ornithine via arginase (
CAR1), followed by ornithine decarboxylation to PUT (
SPE1), and then extended to SPD (
SPE3) and SPM (
SPE4) (
Figure 7).
Although some organisms possess a direct arginine decarboxylation pathway that produces agmatine,
S. cerevisiae lacks the enzymes required for this alternative route, such as arginine decarboxylase and agmatinase [
15]. Therefore, the enhanced polyamine production in 3L63 is most plausibly attributed to the upregulation of the conventional ornithine decarboxylase pathway, facilitated by increased arginine availability and potentially improved flux regulation via
ARG3. Additionally, functional enrichment analysis of the 3L63 variant genes revealed significant GO terms, including metabolic process, primary metabolic process, and regulation of metabolic process, suggesting that the mutation landscape in 3L63 may contribute to broad metabolic rewiring. This finding likely supports the enhanced biosynthetic capacity of polyamines.
Environmental stress also influences amino acid metabolism. Heat or nutrient stress can promote the intracellular accumulation of arginine and other amino acids, thereby supporting stress resistance and cell survival in
S. cerevisiae [
43,
44]. Genetic modifications may render standard culture conditions physiologically stressful and elicit similar metabolic responses.
In summary, our findings support a hypothetical mechanism whereby mutations in key metabolic genes, such as ARG3, result in elevated intracellular arginine levels. This surplus arginine is channeled through the native ornithine decarboxylation pathway, ultimately leading to increased SPD production. These results support the potential of 3L63 as a high-polyamine-producing, non-GMO yeast strain for application in functional foods. Further validation using metabolic flux analysis, transcriptomic profiling, and enzyme activity assays will provide deeper insights into the regulatory networks underlying polyamine overproduction.
4.3. Functional Benefits of SPD: Autophagy and Healthy Aging
SPD is a naturally occurring polyamine that has garnered significant attention owing to its role in promoting autophagy, maintaining cellular homeostasis, and supporting healthy aging [
8,
9,
10,
45]. Autophagy is a highly conserved intracellular recycling process that declines with age and is linked to the pathogenesis of numerous age-related diseases including neurodegeneration, cardiovascular dysfunction, and skin aging. SPD reactivates autophagy through epigenetic and metabolic regulation, thus mimicking the effects of caloric restriction and inducing longevity [
45]. This underlies the broad protective effect observed in preclinical and epidemiological studies [
24].
Although no formal dietary reference intake has been established, a growing body of research derived from both animal and human studies supports a daily intake of approximately 15–30 mg SPD to promote longevity and reduce the risk of age-associated diseases [
8,
46,
47]. For instance, Eisenberg et al. [
8] reported that dietary SPD extended lifespan and improved cardiovascular health in mice, whereas Kiechl et al. [
37] demonstrated inverse associations between SPD intake and mortality, as well as a lower incidence of cardiovascular events, in a human cohort study.
Achieving the longevity-promoting intake range through diet alone is challenging. Wheat germ (~24 mg/100 g), fermented soybeans (natto; ~12–20 mg/100 g), mushrooms (~5–10 mg/100 g), and green peas (~4–6 mg/100 g) are SPD-rich foods [
12,
13] that are typically not consumed in sufficient quantities. The average daily intake in Western and Asian populations is just 10–12 mg [
11,
47]. Our 3L63 yeast-derived product offers a potent and convenient dietary source of SPD, containing over 2.0 mg/g. Only 10 g/d provides approximately 20 mg, which is sufficient to bridge this dietary gap.
In addition to inadequate dietary intake, the body’s endogenous production of SPD declines with age, with blood concentrations averaging 0.63 μM at birth but falling to 0.25 μM by age 70 [
48], a drop linked to cellular dysfunction and accelerated aging. This decline corresponds to ~0.28 mg less SPD in 5 L human blood. At 2.0 mg/g, only ~160 mg of 3L63 yeast powder would theoretically restore this youthful blood concentration. However, normalizing blood levels may not equate to replenishing whole-body stores or sustaining tissue autophagy.
For physiological and longevity benefits, the literature supports a daily intake of 15–30 mg SPD; at 2.0 mg/g, this corresponds to roughly 7.5–15 g of 3L63 powder. A practical target of 10–15 g/d (≈20–30 mg SPD) not only fills the typical dietary gap but also provides a margin for incomplete absorption, first-pass metabolism, and tissue distribution. This distinction between the minimal dose required to restore blood levels and the sustained intake required for systemic effects underscores the advantages of using 3L63 as a functional food ingredient for healthy aging. Notably, in a clinical study among 50–70 year-old men, daily supplementation with 300 mg of 3L63 yeast commercial product (together with 1 g BCAA given to both control and test groups) for 8 weeks led to significant increases in skeletal muscle index (SMI), muscle mass, and fat-free mass [
49]. This suggests that even relatively low doses of 3L63 than theoretical estimation can confer measurable functional benefits in humans.
To further assess the biological relevance of the activity of 3L63-derived SPD, we performed a series of functional assays. Autophagy induction was observed in HeLa cells using a fluorescent assay, where the 3L63 yeast product (containing 0.86 μM SPD) significantly increased autophagy activity by ~130% relative to that observed in the control, exceeding the effect of pure SPD at 100 μM. Similarly, a tfLC3 assay revealed that even low concentrations of the 3L63 product (0.1–5 μM SPD) activated autophagic flux in cells. These results suggest a synergistic effect between SPD and other yeast-derived components, enhancing autophagy beyond that achieved with SPD alone.
Using HDFs, we demonstrated that the 3L63 yeast product promoted cell proliferation in both normal and senescent fibroblasts. Given that autophagy supports proteostasis, mitochondrial function, and resistance to oxidative stress, the enhanced proliferation of aging fibroblasts likely reflects the restoration of these core anti-aging mechanisms. These findings reinforce the role of SPD-induced autophagy as a central mediator of cellular rejuvenation, particularly in skin health, and highlight that 3L63 yeast is a promising candidate for anti-aging intervention via nutritional or topical routes. Additionally, in our previous study using C2C12 myoblasts, we showed that 25 μg/mL yeast-derived SPD promoted myogenic differentiation and enhanced muscle cell viability, indicating a role in supporting muscle growth [
49]. Building on this, SPD-enriched 3L63 yeast may also confer functional benefits for skeletal muscle health—further underscoring its potential as a versatile anti-aging dietary ingredient.
Although our data established both biochemical potency and functional activity, further studies are required to investigate the bioavailability, metabolic fate, and more clinical outcomes of 3L63-derived SPD. Factors such as individual microbiome profiles, gut polyamine metabolism, and potential synergy with other nutrients should be considered in future human trials for optimal formulation development. To address these limitations, future studies should include controlled human trials to assess SPD bioavailability, pharmacokinetics, and clinical outcomes. Metabolomic analyses could elucidate the metabolic fate of SPD and identify individual factors, such as microbiome composition, that affect its efficacy. Additionally, investigating synergistic effects with other nutrients or bioactive compounds could inform optimal formulation strategies for functional foods or supplements. Incorporating these approaches will strengthen the translational relevance of 3L63-derived SPD and guide evidence-based product development.
5. Conclusions
The 3L63 SPD-enriched yeast strain developed in this study offers a novel and scalable approach for addressing the growing demand for natural polyamine supplementation. Owing to its high SPD content, GRAS status, scientific formulation, and bioactivity, it is a promising next-generation ingredient for functional foods, dietary supplements, and healthy aging applications.
6. Patents
Masanori Tamakoshi, Satoko Imaruoka, and Kazuto Ikemoto, Polyamine-rich yeast and food and beverage composition containing the same, Japanese Patent No. 6663598, filed on March 2, 2018.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: Workflow of polyamine derivatization using dansyl chloride for HPLC analysis; Figure S2: Functional enrichment analysis result obtained from g:Profiler; Figure S3: Protein variant modelling and docking simulation; Figure S4: HPLC chromatogram of free amino acid analysis; Figure S5: Autophagy activation effect using western blotting (flux assay); Table S1: The analytical methods employed for nutritional composition of 3L63 yeast-derived powder; Table S2: Safety and quality test results of the 3L63 yeast-derived powder containing 15% α-cyclodextrin; Table S3: 3L63 yeast product specification for quality standard; Table S4: KEGG pathway enrichment analysis results; Table S5: Summary of key SNPs; Table S6: Amino acid and polyamine content in K7 and 3L63 strains after overnight cultivation, and 3L63 yeast derived product.
Author Contributions
Conceptualization, Kazuto Ikemoto; Data curation, Tomoyo Koshizawa, Tomoe Numaguchi and Nur Syafiqah Mohamad Ishak; Formal analysis, Tomoyo Koshizawa, Tomoe Numaguchi and Masanori Tamakoshi; Investigation, Tomoyo Koshizawa, Tomoe Numaguchi, Masanori Tamakoshi and Nur Syafiqah Mohamad Ishak; Project administration, Yuuki Sato and Katsuyuki Hashimoto; Supervision, Yuuki Sato, Katsuyuki Hashimoto and Kazuto Ikemoto; Validation, Nur Syafiqah Mohamad Ishak; Visualization, Tomoyo Koshizawa and Nur Syafiqah Mohamad Ishak; Writing – original draft, Nur Syafiqah Mohamad Ishak and Kazuto Ikemoto; Writing – review & editing, Tomoyo Koshizawa, Tomoe Numaguchi, Nur Syafiqah Mohamad Ishak and Kazuto Ikemoto.
Funding
All research was funded by the Mitsubishi Gas Chemical Company, Inc.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Acknowledgments
We thank the members of the Life Sciences Department and Niigata Research Laboratory of Mitsubishi Gas Chemical Co., Ltd. for their assistance. We would like to thank Editage (
www.editage.jp) for English language editing and their assistance in creating the graphical abstract for this paper.
Conflicts of Interest
All authors are salaried employees of the Mitsubishi Gas Chemical Co., Ltd.
Abbreviations
The following abbreviations are used in this manuscript:
| BP |
biological process |
| CC |
cellular component |
| GMO |
genetically modified organism |
| GO |
Gene Ontology |
| GRAS |
Generally Recognized as Safe |
| HDF |
human dermal fibroblast |
| HPLC |
high-performance liquid chromatography |
| MF |
molecular function |
| PUT |
putrescine |
| SAM |
S-adenosylmethionine |
| SPD |
spermidine |
| SPM |
spermine |
| tfLC3 |
tandem fluorescent-tagged LC3 |
References
- World Health Organization. World Health Statistics 2025: Monitoring Health for the SDGs, Sustainable Development Goals.
- United Nations Department of Economic and Social Affairs, P. D. United Nations Department of Economic and Social Affairs, P. D. World Population Prospects 2024: Summary of Results (UN DESA/POP/2024/TR/NO. 9).
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Rea, I.M.; Gibson, D.S.; McGilligan, V.; McNerlan, S.E.; Alexander, H.D.; Ross, O.A. Age and age-related diseases: role of inflammation triggers and cytokines. Front. Immunol. 2018, 9, 586. [Google Scholar] [CrossRef] [PubMed]
- Mills, K.F.; Yoshida, S.; Stein, L.R.; Grozio, A.; Kubota, S.; Sasaki, Y.; Redpath, P.; Migaud, M.E.; Apte, R.S.; Uchida, K.; et al. Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metab. 2016, 24, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Pyo, I.S.; Yun, S.; Yoon, Y.E.; Choi, J.-W.; Lee, S.-J. Mechanisms of aging and the preventive effects of resveratrol on age-related diseases. Molecules 2020, 25, 4649. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E. Mammalian polyamine metabolism and function. IUBMB Life 2009, 61, 880–894. [Google Scholar] [CrossRef]
- Eisenberg, T.; Abdellatif, M.; Schroeder, S.; Primessnig, U.; Stekovic, S.; Pendl, T.; Harger, A.; Schipke, J.; Zimmermann, A.; Schmidt, A.; et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat. Med. 2016, 22, 1428–1438. [Google Scholar] [CrossRef]
- Niechcial, A.; Schwarzfischer, M.; Wawrzyniak, M.; Atrott, K.; Laimbacher, A.; Morsy, Y.; Katkeviciute, E.; Häfliger, J.; Westermann, P.; Akdis, C.A.; et al. Spermidine ameliorates colitis via induction of anti-inflammatory macrophages and prevention of intestinal dysbiosis. J. Crohns Colitis 2023, 17, 1489–1503. [Google Scholar] [CrossRef]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Büttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef]
- Nishibori, N.; Fujihara, S.; Akatuki, T. Amounts of polyamines in foods in Japan and intake by Japanese. Food Chem. 2007, 100, 491–497. [Google Scholar] [CrossRef]
- Okamoto, A.; Sugi, E.; Koizumi, Y.; Yanagida, F.; Udaka, S. Polyamine content of ordinary foodstuffs and various fermented foods. Biosci. Biotechnol. Biochem. 1997, 61, 1582–1584. [Google Scholar] [CrossRef]
- Atiya Ali, M.; Poortvliet, E.; Strömberg, R.; Yngve, A. Polyamines in foods: development of a food database. Food Nutr. Res. 2011, 55, 5572. [Google Scholar] [CrossRef]
- Muñoz-Esparza, N.C.; Latorre-Moratalla, M.L.; Comas-Basté, O.; Toro-Funes, N.; Veciana-Nogués, M.T.; Vidal-Carou, M.C. Polyamines in food. Front. Nutr. 2019, 6, 108. [Google Scholar] [CrossRef]
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining mysteries of molecular biology: the role of polyamines in the cell. J. Mol. Biol. 2015, 427, 3389–3406. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E. Functions of polyamines in mammals. J. Biol. Chem. 2016, 291, 14904–14912. [Google Scholar] [CrossRef] [PubMed]
- Frewer, L.J.; van der Lans, I.A.; Fischer, A.R.H.; Reinders, M.J.; Menozzi, D.; Zhang, X.; van den Berg, I.; Zimmermann, K.L. Public perceptions of Agri-food applications of genetic modification—A systematic review and meta-analysis. Trends Food Sci. Technol. 2013, 30, 142–152. [Google Scholar] [CrossRef]
- Akao, T.; Yashiro, I.; Hosoyama, A.; Kitagaki, H.; Horikawa, H.; Watanabe, D.; Akada, R.; Ando, Y.; Harashima, S.; Inoue, T.; et al. Whole-genome sequencing of sake yeast Saccharomyces cerevisiae Kyokai No. 7. DNA Res. 2011, 18, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Bagherniya, M.; Butler, A.E.; Barreto, G.E.; Sahebkar, A. The effect of fasting or calorie restriction on autophagy induction: a review of the literature. Ageing Res. Rev. 2018, 47, 183–197. [Google Scholar] [CrossRef]
- Chung, K.W.; Chung, H.Y. The effects of calorie restriction on autophagy: role on aging intervention. Nutrients 2019, 11, 2923. [Google Scholar] [CrossRef]
- Madeo, F.; Zimmermann, A.; Maiuri, M.C.; Kroemer, G. Essential role for autophagy in life span extension. J. Clin. Invest. 2015, 125, 85–93. [Google Scholar] [CrossRef]
- Eckhart, L.; Tschachler, E.; Gruber, F. Autophagic control of skin aging. Front Cell Dev Biol 2019, 7. [Google Scholar] [CrossRef]
- Klapan, K.; Simon, D.; Karaulov, A.; Gomzikova, M.; Rizvanov, A.; Yousefi, S.; Simon, H.U. Autophagy and skin diseases. Front Pharmacol 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Madeo, F.; Eisenberg, T.; Pietrocola, F.; Kroemer, G. Spermidine in health and disease. Science (1979) 2018, 359. [Google Scholar] [CrossRef] [PubMed]
- Schibalski, R.S.; Shulha, A.S.; Tsao, B.P.; Palygin, O.; Ilatovskaya, D. V. The role of polyamine metabolism in cellular function and physiology. American Journal of Physiology-Cell Physiology 2024, 327, C341–C356. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g:Profiler—interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 2022, 31, 47–53. [Google Scholar] [CrossRef]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev 2015, 82, 518–529. [Google Scholar] [CrossRef]
- Kimura, S.; Noda, T.; Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007, 3, 452–460. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.-X.; Cao, Y. CB-Dock2: Improved protein–ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res 2022, 50, W159–W164. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration (FDA) Code of Federal Regulations Title 21, §184.1983—Baker’s Yeast.
- Akao, T. The genome analysis of sake Yeast Kyokai No. 7 and foresights of sake yeast genomics. J. Brew. Soc. Jpn. 2012, 107, 366–380. [Google Scholar] [CrossRef]
- Akao, T. Progress in the genomics and genome-wide study of sake yeast. Biosci. Biotechnol. Biochem. 2019, 83, 1463–1472. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, T.; Yamada, K.; Nagahisa, K.; Dinh, T.N.; Furusawa, C.; Katakura, Y.; Shioya, S.; Shimizu, H. Proteomic analysis of responses to osmotic stress in laboratory and sake-brewing strains of Saccharomyces cerevisiae. Process Biochem. 2009, 44, 647–653. [Google Scholar] [CrossRef]
- Watanabe, D.; Kajihara, T.; Sugimoto, Y.; Takagi, K.; Mizuno, M.; Zhou, Y.; Chen, J.; Takeda, K.; Tatebe, H.; Shiozaki, K.; et al. Nutrient signaling via the TORC1-Greatwall-PP2AB55δ pathway is responsible for the high initial rates of alcoholic fermentation in sake yeast strains of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2019, 85, e02083–18. [Google Scholar] [CrossRef]
- Kim, S.K.; Jo, J.H.; Park, Y.C.; Jin, Y.S.; Seo, J.H. Metabolic engineering of Saccharomyces cerevisiae for production of spermidine under optimal culture conditions. Enzyme Microb Technol 2017, 101, 30–35. [Google Scholar] [CrossRef]
- Qin, J.; Krivoruchko, A.; Ji, B.; Chen, Y.; Kristensen, M.; Özdemir, E.; Keasling, J.D.; Jensen, M.K.; Nielsen, J. Engineering yeast metabolism for the discovery and production of polyamines and polyamine Analogues. Nat Catal 2021, 4, 498–509. [Google Scholar] [CrossRef]
- Matencio, A.; Navarro-Orcajada, S.; García-Carmona, F.; López-Nicolás, J.M. Applications of cyclodextrins in food science. A Review. Trends Food Sci Technol 2020, 104, 132–143. [Google Scholar] [CrossRef]
- Astray, G.; Gonzalez-Barreiro, C.; Mejuto, J.C.; Rial-Otero, R.; Simal-Gándara, J. A review on the use of cyclodextrins in foods. Food Hydrocoll. 2009, 23, 1631–1640. [Google Scholar] [CrossRef]
- Crabeel, M.; Messenguy, F.; Lacroute, F.; Glansdorff, N. Cloning arg3, the gene for ornithine carbamoyltransferase from Saccharomyces cerevisiae: expression in Escherichia coli requires secondary mutations; production of plasmid beta-lactamase in yeast. Proc. Natl. Acad. Sci. USA. 1981, 78, 5026–5030. [Google Scholar] [CrossRef]
- Messenguy, F. Regulation of arginine biosynthesis in Saccharomyces cerevisiae: isolation of a cis-dominant, constitutive mutant for ornithine carbamoyltransferase synthesis. J. Bacteriol. 1976, 128, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Wiedemann, N.; Kammerer, B. Heat stress-induced metabolic remodeling in Saccharomyces cerevisiae. Metabolites 2019, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Kawahata, M.; Masaki, K.; Fujii, T.; Iefuji, H. Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res. 2006, 6, 924–936. [Google Scholar] [CrossRef]
- Madeo, F.; Eisenberg, T.; Büttner, S.; Ruckenstuhl, C.; Kroemer, G. Spermidine: A novel autophagy inducer and longevity elixir. Autophagy 2010, 6, 160–162. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, S.; Hofer, S.J.; Zimmermann, A.; Pechlaner, R.; Dammbrueck, C.; Pendl, T.; Marcello, G.M.; Pogatschnigg, V.; Bergmann, M.; Müller, M.; et al. Dietary spermidine improves cognitive function. Cell Rep. 2021, 35, 108985. [Google Scholar] [CrossRef] [PubMed]
- Kiechl, S.; Pechlaner, R.; Willeit, P.; Notdurfter, M.; Paulweber, B.; Willeit, K.; Werner, P.; Ruckenstuhl, C.; Iglseder, B.; Weger, S.; et al. Higher spermidine intake is linked to lower mortality: A prospective population-based study. Am. J. Clin. Nutr. 2018, 108, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Rudman, D.; Kutner, M.H.; Chawla, R.K.; Goldsmith, M.A.; Blackston, R.D.; Bain, R. Serum and urine polyamines in normal and in short children. J. Clin. Invest. 1979, 64, 1661–1668. [Google Scholar] [CrossRef]
- Tomoe, Numaguchi; Yuuto, Takenaka; Yuuki Sato <i>Food Style 21 Vol. 10</i>. <b>2023</b>, pp. Tomoe Numaguchi; Yuuto Takenaka; Yuuki Sato Food Style 21 Vol. 10. 2023, pp. 45–48.
Figure 1.
Scale-up of spermidine-producing yeast fermentation and product preparation. (a) Microscopic images (bright field and fluorescence) showing the morphology and viability of the engineered yeast strain. (b) Fermentation was performed at increasing scales using different fermentation vessels: shake flasks, 3-L jar fermenter, and 30-L jar fermenter. (c) Cultured yeast biomass was dried and processed into powder and tablet forms (d) Chemical structure of the major polyamines produced at high levels in this yeast strain: putrescine (PUT), spermidine (SPD), and spermine (SPM), in which SPD was the most abundance.
Figure 1.
Scale-up of spermidine-producing yeast fermentation and product preparation. (a) Microscopic images (bright field and fluorescence) showing the morphology and viability of the engineered yeast strain. (b) Fermentation was performed at increasing scales using different fermentation vessels: shake flasks, 3-L jar fermenter, and 30-L jar fermenter. (c) Cultured yeast biomass was dried and processed into powder and tablet forms (d) Chemical structure of the major polyamines produced at high levels in this yeast strain: putrescine (PUT), spermidine (SPD), and spermine (SPM), in which SPD was the most abundance.
Figure 2.
Comparison of polyamine content in different yeast strains and foods. (a) HPLC chromatograms of parent K7 (top) and 3L63 yeast strains (bottom). (b) Amounts of PUT, SPD, and SPM (μmol/g dry weight) were compared between the parent K7 strain, newly developed strain 3L63, commercial 3L63 yeast product co-formulated with α-cyclodextrin, and representative foods such as natto and mushroom. The 3L63 strain exhibited the highest total polyamine content, particularly SPD, suggesting its potential as a dietary source of polyamines. N.D.: Not detected.
Figure 2.
Comparison of polyamine content in different yeast strains and foods. (a) HPLC chromatograms of parent K7 (top) and 3L63 yeast strains (bottom). (b) Amounts of PUT, SPD, and SPM (μmol/g dry weight) were compared between the parent K7 strain, newly developed strain 3L63, commercial 3L63 yeast product co-formulated with α-cyclodextrin, and representative foods such as natto and mushroom. The 3L63 strain exhibited the highest total polyamine content, particularly SPD, suggesting its potential as a dietary source of polyamines. N.D.: Not detected.
Figure 3.
Functional enrichment analysis of genes with high- and moderate-impact mutations in 3L63 yeast strain. Bubble plot visualization of Gene Ontology (GO), and pathway enrichment results obtained from g:Profiler using the filtered gene list (n = 860). GO terms are categorized into molecular function (GO:MF, red), biological process (GO:BP, orange), and cellular component (GO:CC, green), KEGG pathway (pink) and transcription factors (TF, blue). The top significantly enriched biological processes include metabolic process (GO:0008152, P
adj = 1.070 × 10
-4), primary metabolic process (GO:0044238, P
adj = 3.866 × 10
-3), and regulation of metabolic process (GO:0019222, Padj = 6.171 × 10
-4). The dotted line indicates the –log
10(
p-adjusted) threshold. Detailed results are presented in Supplementary information (
Figure S2).
Figure 3.
Functional enrichment analysis of genes with high- and moderate-impact mutations in 3L63 yeast strain. Bubble plot visualization of Gene Ontology (GO), and pathway enrichment results obtained from g:Profiler using the filtered gene list (n = 860). GO terms are categorized into molecular function (GO:MF, red), biological process (GO:BP, orange), and cellular component (GO:CC, green), KEGG pathway (pink) and transcription factors (TF, blue). The top significantly enriched biological processes include metabolic process (GO:0008152, P
adj = 1.070 × 10
-4), primary metabolic process (GO:0044238, P
adj = 3.866 × 10
-3), and regulation of metabolic process (GO:0019222, Padj = 6.171 × 10
-4). The dotted line indicates the –log
10(
p-adjusted) threshold. Detailed results are presented in Supplementary information (
Figure S2).
Figure 4.
Effects of 3L63 yeast product on proliferation of human dermal fibroblasts (HDFs) in both (a) normal and (b) H2O2-induced senescent populations. The amount of 3L63 yeast added for the treatment was adjusted to achieve concentration of 12.5, 25, 50. and 100 μg/mL yeasts. The data indicate mean ± SE (n = 8). Asterisks denote a significant difference according to paired t-test, (*p < 0.05) when compared with control cells. Hashtags denote a significant difference according to paired t-test, (#p < 0.05) compared with H2O2-induced control.
Figure 4.
Effects of 3L63 yeast product on proliferation of human dermal fibroblasts (HDFs) in both (a) normal and (b) H2O2-induced senescent populations. The amount of 3L63 yeast added for the treatment was adjusted to achieve concentration of 12.5, 25, 50. and 100 μg/mL yeasts. The data indicate mean ± SE (n = 8). Asterisks denote a significant difference according to paired t-test, (*p < 0.05) when compared with control cells. Hashtags denote a significant difference according to paired t-test, (#p < 0.05) compared with H2O2-induced control.
Figure 5.
Autophagy induction assays using CTYO-ID dye. (a) Representative images of CTYO-ID (autophagy activity) and Heochst 33342 (nuclei) staining in HeLa cells. The cells were treated with 50 μg/L 3L63 containing 0.86 μM SPD, 100 μM pure SPD, and 0.86 uM pure SPD, and autophagy activity was compared with that of untreated control. (b) Fluorescent intensity (FI) was quantified using fluorescence imaging analysis. The data indicate mean ± SE (n = 8). Different letters above bars idnicate significant differences among treatment conditions (one-way ANOVA, all pairwise multiple comparisons with the Tukey’s HSD test, p < 0.05). .
Figure 5.
Autophagy induction assays using CTYO-ID dye. (a) Representative images of CTYO-ID (autophagy activity) and Heochst 33342 (nuclei) staining in HeLa cells. The cells were treated with 50 μg/L 3L63 containing 0.86 μM SPD, 100 μM pure SPD, and 0.86 uM pure SPD, and autophagy activity was compared with that of untreated control. (b) Fluorescent intensity (FI) was quantified using fluorescence imaging analysis. The data indicate mean ± SE (n = 8). Different letters above bars idnicate significant differences among treatment conditions (one-way ANOVA, all pairwise multiple comparisons with the Tukey’s HSD test, p < 0.05). .
Figure 6.
Analysis of autophagy using HeLa cells transfected with tfLC3 construct. (a) Fluorescent images showing the localization of LC3 puncta in autophagic structures for control and SPD-enriched 3L63 yeast product. mRFP-LC3 (magenta) stably marks all autophagic structures including autophagosomes and autolysosomes. GFP-LC3 (green) marks early autophagic structures (isolation membranes and autophagosomes), but signal is quenched in acidic autolysosomes. DAPI (blue) allows nuclear staining for cell visualization. (b) Quantification of fluorescent intensity based on GFP/RFP ratio using fluorescence imaging analysis. The data indicate mean ± SE (n = 3). Asterisks denote a significant difference according to paired t-test, (**p < 0.01; ***p < 0.001) when compared with control cells.
Figure 6.
Analysis of autophagy using HeLa cells transfected with tfLC3 construct. (a) Fluorescent images showing the localization of LC3 puncta in autophagic structures for control and SPD-enriched 3L63 yeast product. mRFP-LC3 (magenta) stably marks all autophagic structures including autophagosomes and autolysosomes. GFP-LC3 (green) marks early autophagic structures (isolation membranes and autophagosomes), but signal is quenched in acidic autolysosomes. DAPI (blue) allows nuclear staining for cell visualization. (b) Quantification of fluorescent intensity based on GFP/RFP ratio using fluorescence imaging analysis. The data indicate mean ± SE (n = 3). Asterisks denote a significant difference according to paired t-test, (**p < 0.01; ***p < 0.001) when compared with control cells.
Figure 7.
Structural and metabolic integration of arginine and polyamine biosynthesis in
S. cerevisiae. The to the left depicts the molecular surface of two enzyme models of ARG3 (K7 and 3L63), showing predicted ligand-binding cavities in red, visualized using molecular docking simulations. Predicted cavity volume (ų) and binding affinity (AutodockVina score) are annotated to indicate differences in substrate accommodation or enzymatic potential between variants. The image to the right illustrates the metabolic interplay between arginine biosynthesis and polyamine biosynthesis, based on the
S. cerevisiae KEGG pathway [
27]. Arrows indicate enzymatic reactions connecting these two pathways, highlighting steps that may influence intracellular SPD levels.
Figure 7.
Structural and metabolic integration of arginine and polyamine biosynthesis in
S. cerevisiae. The to the left depicts the molecular surface of two enzyme models of ARG3 (K7 and 3L63), showing predicted ligand-binding cavities in red, visualized using molecular docking simulations. Predicted cavity volume (ų) and binding affinity (AutodockVina score) are annotated to indicate differences in substrate accommodation or enzymatic potential between variants. The image to the right illustrates the metabolic interplay between arginine biosynthesis and polyamine biosynthesis, based on the
S. cerevisiae KEGG pathway [
27]. Arrows indicate enzymatic reactions connecting these two pathways, highlighting steps that may influence intracellular SPD levels.
Table 1.
Nutritional composition of 3L63 yeast powder (per 100 g).
Table 1.
Nutritional composition of 3L63 yeast powder (per 100 g).
| Item |
Measured value |
| Moisture |
4.5 g |
Protein*
(excluding Polyamines) |
45.7 g
(45.1 g) |
| Fat |
10.0 g |
| Carbohydrate |
29.9 g |
| Ash |
9.9 g |
| Energy |
392 kcal |
| Sodium |
25 mg |
| Salt Equivalent |
0.064 g |
Table 2.
Polyamine content in dried yeast harvested from 3L63 and K7 strains.
Table 2.
Polyamine content in dried yeast harvested from 3L63 and K7 strains.
| Strain |
Putrescine (mg/g) |
Spermidine (mg/g) |
Spermine (mg/g) |
Total (mg/g) |
| K7 |
N.D. |
0.51 |
0.45 |
0.96 |
| 3L63 |
1.07 |
4.24 |
0.36 |
5.66 |
| Commercial product * |
0.63 |
2.26 |
0.19 |
3.08 |
Table 3.
Selected free amino acid and polyamine data for S. cerevisiae K7 and 3L63 strain in flask cultivation.
Table 3.
Selected free amino acid and polyamine data for S. cerevisiae K7 and 3L63 strain in flask cultivation.
| Amino acid |
K7 strain
(μmol/ g) |
3L63 strain
(μmol/ g) |
| Aspartic acid (Asp) |
4.96 (1.6%) |
5.59 (2.0%) |
| Glutamic acid (Glu) |
134 (43.8%) |
104 (36.7%) |
| Glycine (Gly) |
10.4 (3.4%) |
13.8 (4.9%) |
| Threonine (Thr) |
9.39 (3.1%) |
7.57 (2.7%) |
| Arginine (Arg) |
17.4 (5.7%) |
28.3 (10.0%) |
| Serine (Ser) |
43.8 (14.3%) |
43.5 (15.4%) |
| Alanine (Ala) |
14.9 (4.9%) |
10.5 (3.7%) |
| Valine (Val) |
2.18 (0.7%) |
1.83 (1.83%) |
| Leucine (Leu) |
2.21 (0.7%) |
1.90 (0.7%) |
| Lysine (Lys) |
6.10 (2.0%) |
12.9 (4.6%) |
| Proline (Pro) |
4.91 (1.6%) |
2.21 (0.8%) |
| Total amino acids |
306 (100%) |
283 (100%) |
| Polyamine |
K7 strain
(mg/g)
|
3L63 strain
(mg/g)
|
| Putrescine (Put) |
0.04 |
0.39 |
| Spermidine (Spd) |
1.62 |
3.41 |
| Spermine (Spm) |
0.41 |
0.37 |
| Total polyamines |
2.07 |
4.17 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).