Submitted:
14 August 2025
Posted:
14 August 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sampling and Fungal Isolation
2.2. Morphological Identification of Fungal Population
2.3. Molecular Identification of Fungal Genera
2.4. Phylogenetic Analysis
2.5. Pathogenicity Test of Isolated Fungi

2.6. Isolation Frequency Analysis of Fungal Isolates
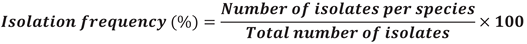
2.7. Data Process and Analysis
3. Results
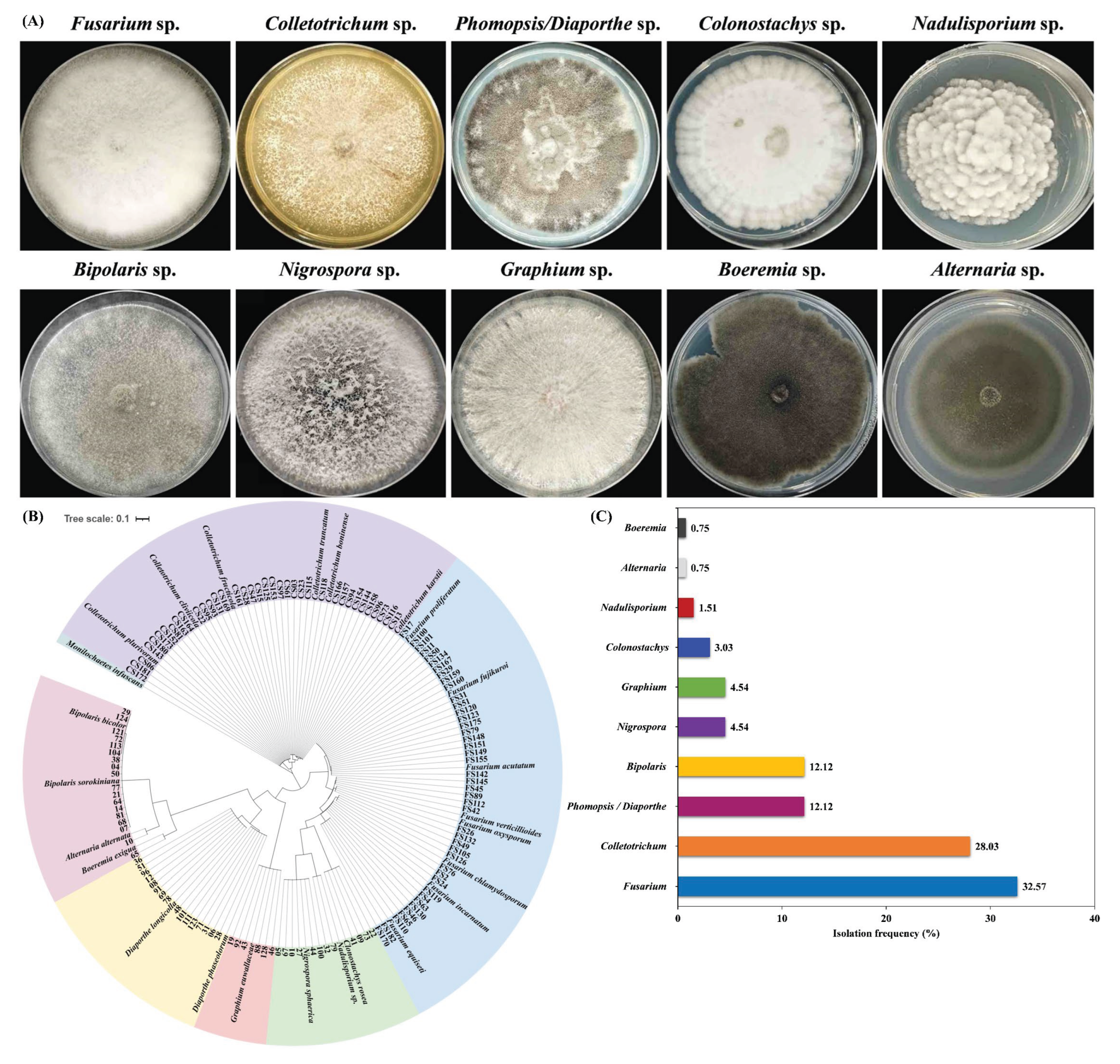
3.1. Identification of Fungal Species Associated with Intercropped Soybean Pods
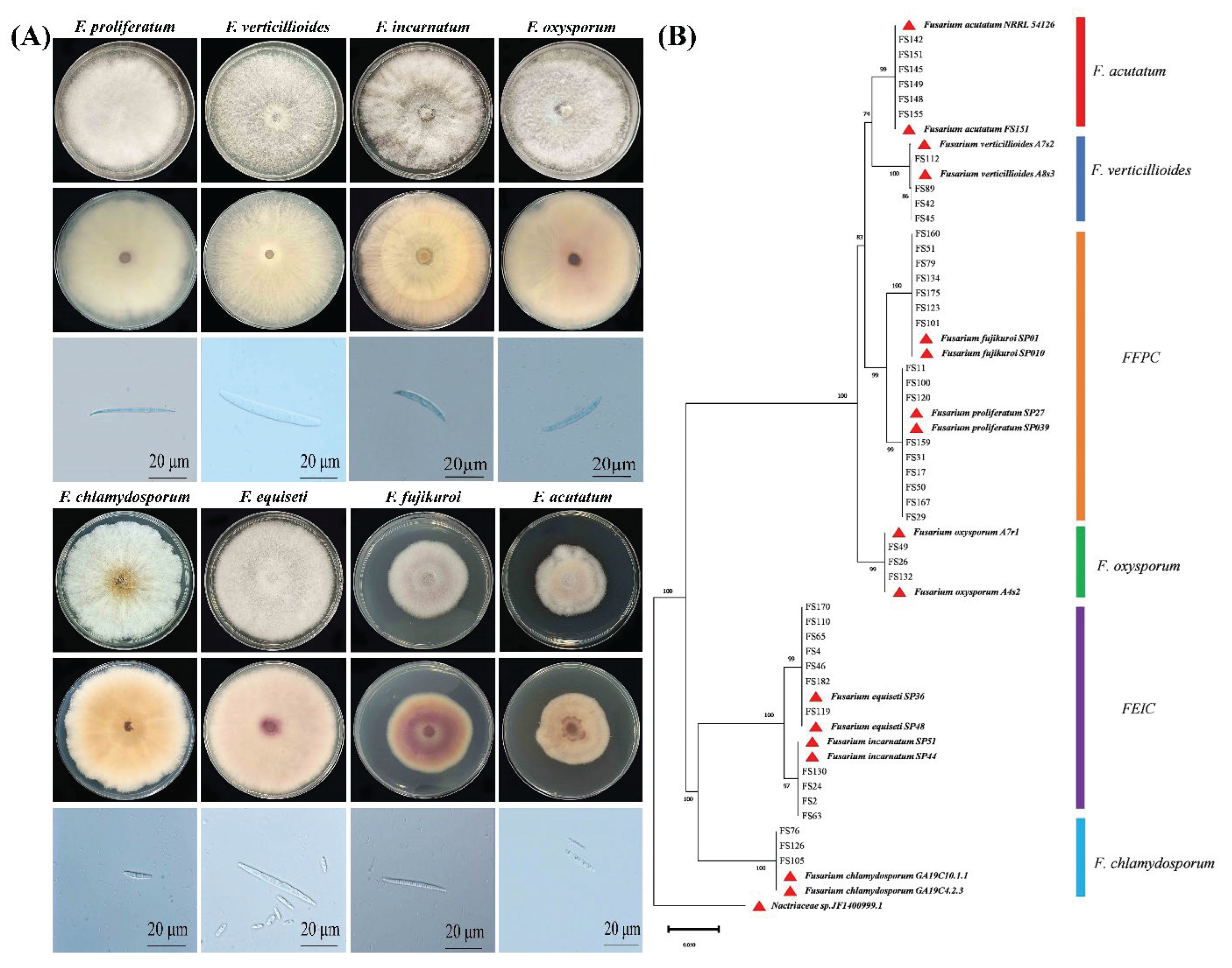
3.2. Identification of Fusarium Species Associated with Soybean Pods
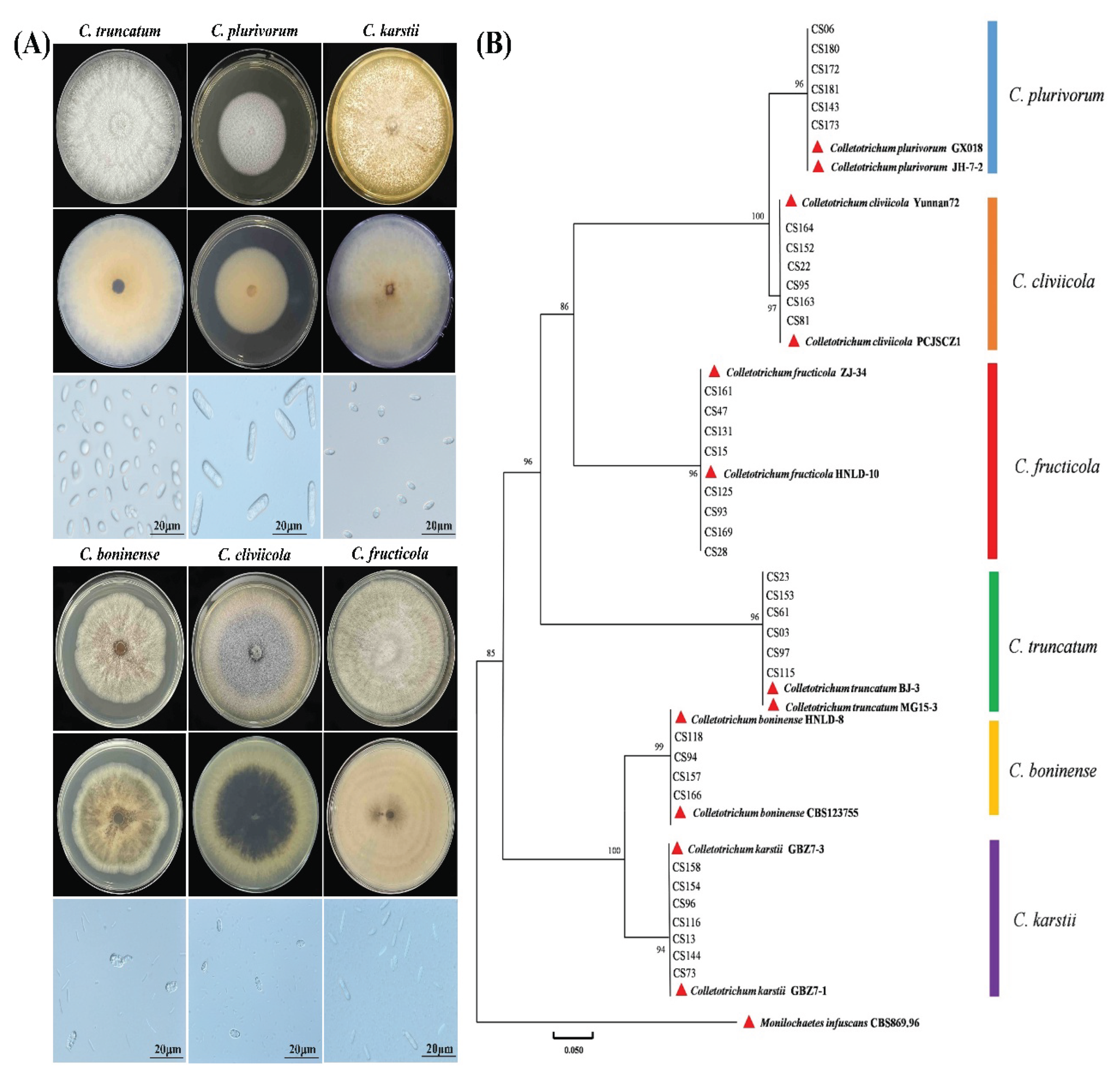
3.3. Identification of Colletotrichum Species Associated with Soybean Pods
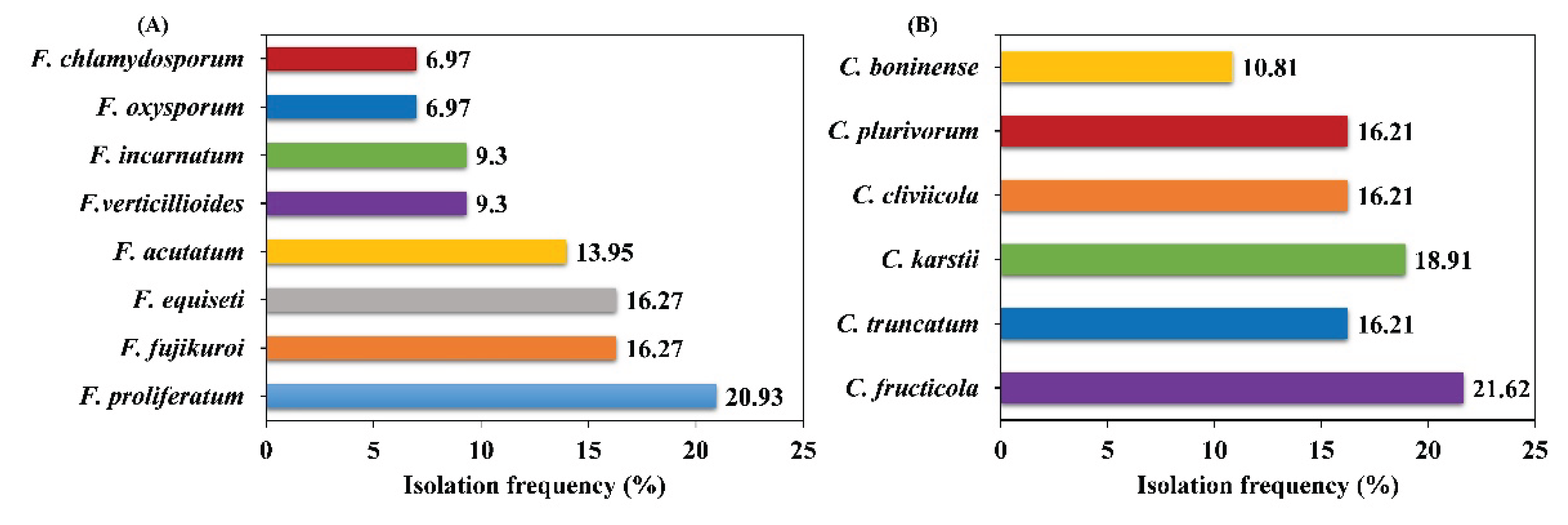
3.4. Isolation Frequency of Fusarium and Colletotrichum Species
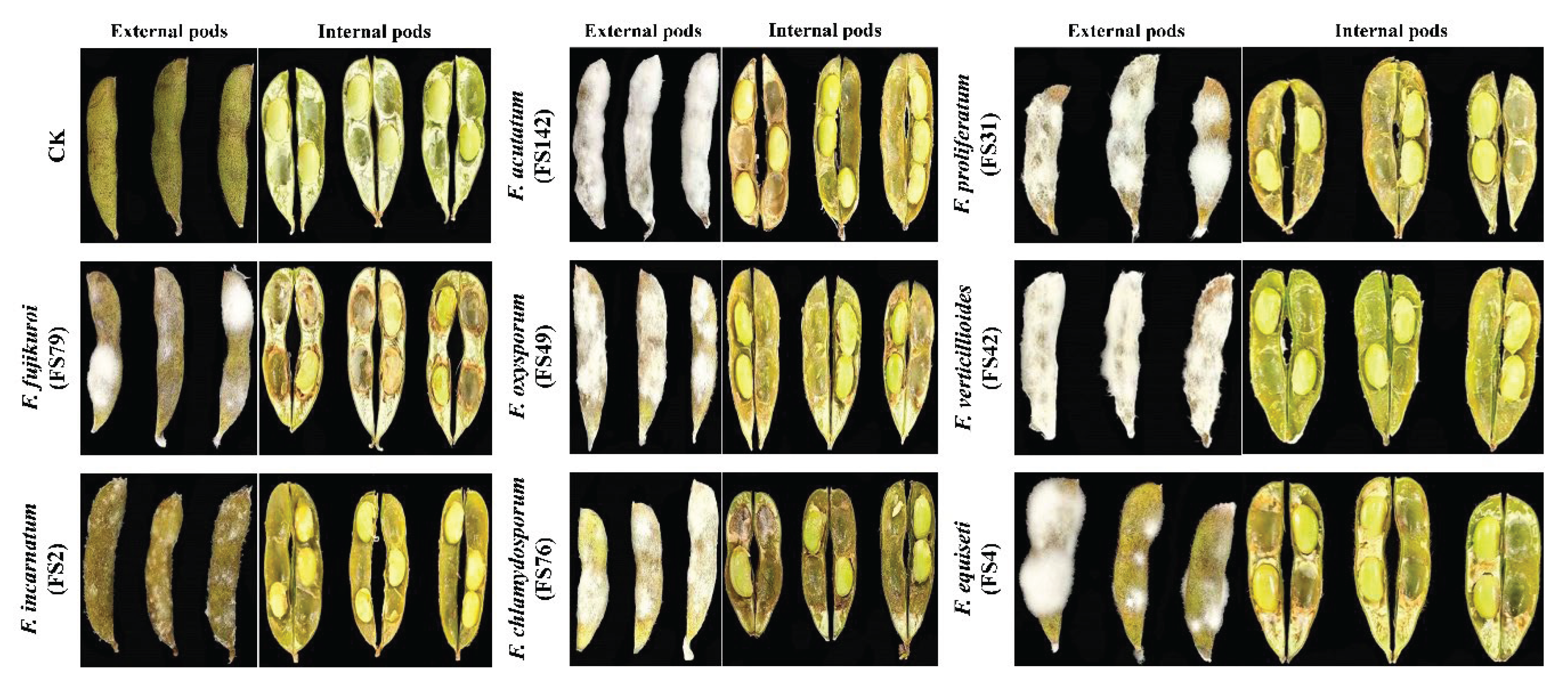
3.5. Pathogenicity of Fusarium Species on Soybean Pods and Seeds
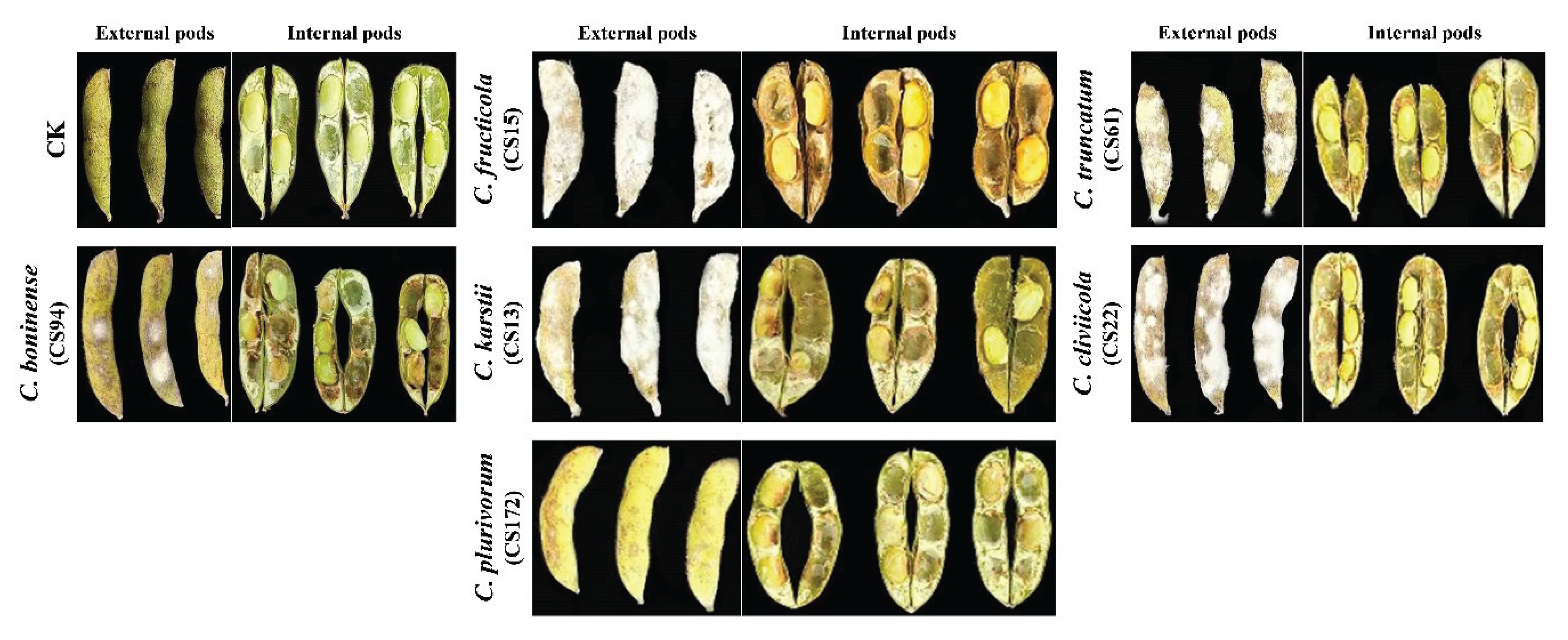
3.6. Pathogenicity of Colletotrichum Species on Soybean Pods and Seeds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Loganathan, M.; Maruthasalam, S.; Shiu, L.Y.; Lien, W.C.; Hsu, W.H.; Lee, P.F.; Yu, C.W.; Lin, C.H. Regeneration of soybean (Glycine max L. Merrill) through direct somatic embryogenesis from the immature embryonic shoot tip. In Vitro Cellular & Developmental Biology-Plant 2010, 46, 265–273. [Google Scholar]
- Kumari, S.; Dambale, A.S.; Samantara, R.; Jincy, M.; Bains, G. Introduction, history, geographical distribution, importance, and uses of soybean (Glycine max L.). In Soybean Production Technology: Physiology, Production and Processing; Springer: 2025; pp. 1-17.
- Modgil, R.; Tanwar, B.; Goyal, A.; Kumar, V. Soybean (Glycine max). In Oilseeds: health attributes and food applications; Springer: 2020; pp. 1-46.
- Belewu, M.; Belewu, K. Comparative physico-chemical evaluation of tiger-nut, soybean and coconut milk sources. International Journal of Agriculture and Biology 2007, 5, e787. [Google Scholar]
- Pagano, M.C.; Miransari, M. The importance of soybean production worldwide. In Abiotic and biotic stresses in soybean production; Elsevier: 2016; pp. 1-26.
- Mishra, R.; Tripathi, M.; Sikarwar, R.; Singh, Y.; Tripathi, N. Soybean (Glycine max L. Merrill): A multipurpose legume shaping our world. Plant Cell Biotechnol. Mol. Biol 2024, 25, 17–37. [Google Scholar] [CrossRef]
- Hosseini, B.; Voegele, R.T.; Link, T.I. Diagnosis of soybean diseases caused by fungal and oomycete pathogens: Existing methods and new developments. Journal of Fungi 2023, 9, 587. [Google Scholar] [CrossRef] [PubMed]
- Naeem, M.; Li, H.; Yan, L.; Raza, M.A.; Gong, G.; Chen, H.; Yang, C.; Zhang, M.; Shang, J.; Liu, T. Characterization and pathogenicity of Fusarium species associated with soybean pods in maize/soybean strip intercropping. Pathogens 2019, 8, 245. [Google Scholar] [CrossRef]
- Arias, M.M.D.; Leandro, L.F.; Munkvold, G.P. Aggressiveness of Fusarium species and impact of root infection on growth and yield of soybeans. Phytopathology 2013, 103, 822–832. [Google Scholar] [CrossRef]
- Barros, G.G.; Zanon, M.S.A.; Chiotta, M.L.; Reynoso, M.M.; Scandiani, M.M.; Chulze, S.N. Pathogenicity of phylogenetic species in the Fusarium graminearum complex on soybean seedlings in Argentina. European Journal of Plant Pathology 2014, 138, 215–222. [Google Scholar] [CrossRef]
- Chang, X.; Li, H.; Naeem, M.; Wu, X.; Yong, T.; Song, C.; Liu, T.; Chen, W.; Yang, W. Diversity of the seedborne fungi and pathogenicity of Fusarium species associated with intercropped soybean. Pathogens 2020, 9, 531. [Google Scholar] [CrossRef]
- Zhang, J.; Xue, A.; Cober, E.; Morrison, M.; Zhang, H.; Zhang, S.; Gregorich, E. Prevalence, pathogenicity and cultivar resistance of Fusarium and Rhizoctonia species causing soybean root rot. Canadian Journal of Plant Science 2013, 93, 221–236. [Google Scholar] [CrossRef]
- Pedrozo, R.; Little, C.R. Fusarium verticillioides inoculum potential influences soybean seed quality. European Journal of Plant Pathology 2017, 148, 749–754. [Google Scholar] [CrossRef]
- Pioli, R.; Mozzoni, L.; Morandi, E. First report of pathogenic association between Fusarium graminearum and soybean. Plant Disease 2004, 88, 220–220. [Google Scholar] [CrossRef] [PubMed]
- Leslie, J.F.; Summerell, B.A. The Fusarium laboratory manual; John Wiley & Sons: 2008.
- Chiotta, M.L.; Alaniz Zanon, M.S.; Palazzini, J.M.; Scandiani, M.M.; Formento, Á.N.; Barros, G.G.; Chulze, S.N. Pathogenicity of Fusarium graminearum and F. meridionale on soybean pod blight and trichothecene accumulation. Plant Pathology 2016, 65, 1492–1497. [Google Scholar] [CrossRef]
- Olszewski, J.; Dzienis, G.; Okorski, A.; Goś, W.; Pszczółkowska, A. Fungal Colonization of the Anatomical Parts of Soybean Seeds Supplied with Different Nitrogen Rates and Inoculated with Bradyrhizobium japonicum. Agriculture 2025, 15, 857. [Google Scholar] [CrossRef]
- Zhang, M.; Shi, Z.; Chen, G.; Cao, A.; Wang, Q.; Yan, D.; Fang, W.; Li, Y. Detection and identification methods and control techniques for crop seed diseases. Agriculture 2023, 13, 1786. [Google Scholar] [CrossRef]
- Mancini, V.; Murolo, S.; Romanazzi, G. Diagnostic methods for detecting fungal pathogens on vegetable seeds. Plant Pathology 2016, 65, 691–703. [Google Scholar] [CrossRef]
- Panwar, S.; Duggirala, K.S.; Yadav, P.; Debnath, N.; Yadav, A.K.; Kumar, A. Advanced diagnostic methods for identification of bacterial foodborne pathogens: Contemporary and upcoming challenges. Critical Reviews in Biotechnology 2023, 43, 982–1000. [Google Scholar] [CrossRef]
- Echarte, L.; Della Maggiora, A.; Cerrudo, D.; Gonzalez, V.; Abbate, P.; Cerrudo, A.; Sadras, V.; Calvino, P. Yield response to plant density of maize and sunflower intercropped with soybean. Field Crops Research 2011, 121, 423–429. [Google Scholar] [CrossRef]
- Abdelmagid, A.; Hafez, M.; Soliman, A.; Adam, L.R.; Daayf, F. First report of Fusarium sporotrichioides causing root rot of soybean in Canada and detection of the pathogen in host tissues by PCR. Canadian Journal of Plant Pathology 2021, 43, 527–536. [Google Scholar] [CrossRef]
- Chang, K.; Hwang, S.; Conner, R.; Ahmed, H.; Zhou, Q.; Turnbull, G.; Strelkov, S.; McLaren, D.; Gossen, B. First report of Fusarium proliferatum causing root rot in soybean (Glycine max L.) in Canada. Crop Protection 2015, 67, 52–58. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, N.; Chang, K.-F.; Hwang, S.-F.; Strelkov, S.E.; Conner, R.L.; McLaren, D.L.; Fu, H.; Harding, M.W.; Turnbull, G.D. Genetic diversity and aggressiveness of Fusarium species isolated from soybean in Alberta, Canada. Crop Protection 2018, 105, 49–58. [Google Scholar] [CrossRef]
- Chang, X.; Dai, H.; Wang, D.; Zhou, H.; He, W.; Fu, Y.; Ibrahim, F.; Zhou, Y.; Gong, G.; Shang, J. Identification of Fusarium species associated with soybean root rot in Sichuan Province, China. European Journal of Plant Pathology 2018, 151, 563–577. [Google Scholar] [CrossRef]
- Yang, X.; Feng, F. Ranges and diversity of soybean fungal diseases in North America. Phytopathology 2001, 91, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wei, X.; Zheng, T.; Gou, Y.N.; Wang, J.; Deng, J.X.; Li, M.J. Evaluation of pathogenic Fusarium spp. associated with soybean seed (Glycine max) in Hubei Province, China. Plant Disease 2022, 106, 3178–3186. [Google Scholar] [CrossRef] [PubMed]
- Abdelmagid1; 2, A.; Hafez1; 3, M.; Lawley, Y.; Adam, L.; Daayf, F. First report of Fusarium cerealis causing root rot on soybean. 2018.
- Ellis, M.; Arias, M.D.; Jimenez, D.C.; Munkvold, G.; Leandro, L. First report of Fusarium commune causing damping-off, seed rot, and seedling root rot on soybean (Glycine max) in the United States. Plant Disease 2013, 97, 284–284. [Google Scholar] [CrossRef]
- Olszak-Przybyś, H.; Korbecka-Glinka, G.; Patkowska, E. Identification and pathogenicity of Fusarium isolated from soybean in Poland. Pathogens 2023, 12, 1162. [Google Scholar] [CrossRef]
- Chang, X.; Naeem, M.; Li, H.; Yan, L.; Liu, T.; Liu, B.; Zhang, H.; Khaskheli, M.; Gong, G.; Zhang, M. First report of Fusarium asiaticum as a causal agent for seed decay of soybean (Glycine max) in Sichuan, China. Plant Disease 2020, 104, 1542–1542. [Google Scholar] [CrossRef]
- Chang, X.; Yan, L.; Naeem, M.; Khaskheli, M.I.; Zhang, H.; Gong, G.; Zhang, M.; Song, C.; Yang, W.; Liu, T. Maize/soybean relay strip intercropping reduces the occurrence of Fusarium root rot and changes the diversity of the pathogenic Fusarium species. Pathogens 2020, 9, 211. [Google Scholar] [CrossRef]
- Arias, M.D.; Munkvold, G.; Ellis, M.; Leandro, L. Distribution and frequency of Fusarium species associated with soybean roots in Iowa. Plant disease 2013, 97, 1557–1562. [Google Scholar] [CrossRef]
- Cannon, P.; Damm, U.; Johnston, P.; Weir, B. Colletotrichum: current status and future directions. Studies in mycology 2012, 73, 181–213. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J. The Top 10 fungal pathogens in molecular plant pathology. Molecular plant pathology 2012, 13, 414–430. [Google Scholar] [CrossRef]
- da Silva, L.L.; Moreno, H.L.A.; Correia, H.L.N.; Santana, M.F.; de Queiroz, M.V. Colletotrichum: species complexes, lifestyle, and peculiarities of some sources of genetic variability. Applied microbiology and biotechnology 2020, 104, 1891–1904. [Google Scholar] [CrossRef]
- Sharma, S.; Gupta, G.; Ramteke, R. Colletotrichum truncatum [(Schw.) Andrus & WD Moore], the causal agent of anthracnose of soybean [Glycine max (L.) Merrill]—A Review. Soybean Res 2011, 9, 31–52. [Google Scholar]
- Yang, H.-C.; Hartman, G.L. Methods and evaluation of soybean genotypes for resistance to Colletotrichum truncatum. Plant disease 2015, 99, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Hartman, G.L.; Rupe, J.C.; Sikora, E.J.; Domier, L.L.; Davis, J.A.; Steffey, K.L. Compendium of soybean diseases and pests; American Phytopathological Society, St. Paul, MN: 2015.
- Cai, L.; Hyde, K.; Taylor, P.; Weir, B.; Waller, J.; Abang, M.; Zhang, J.; Yang, Y.; Phoulivong, S.; Liu, Z. A polyphasic approach for studying Colletotrichum. Fungal Diversity 2009, 39, 183–204. [Google Scholar]
- Damm, U.; Woudenberg, J.; Cannon, P.; Crous, P. Colletotrichum species with curved conidia from herbaceous hosts. Fungal Diversity 2009, 39, 45. [Google Scholar]
- Hyde, K.; Cai, L.; Cannon, P.; Crouch, J.; Crous, P.; Damm, U.; Goodwin, P.; Chen, H.; Johnston, P.; Jones, E. Colletotrichum—names in current use. Fungal Diversity 2009, 39, 147–182. [Google Scholar]
- Yang, H.-C.; Haudenshield, J.; Hartman, G. First report of Colletotrichum chlorophyti causing soybean anthracnose. Plant Disease 2012, 96, 1699–1699. [Google Scholar] [CrossRef]
- Yang, H.-C.; Haudenshield, J.S.; Hartman, G.L. Colletotrichum incanum sp. nov., a curved-conidial species causing soybean anthracnose in USA. Mycologia 2014, 106, 32–42. [Google Scholar] [CrossRef]
- Khakimov, A.; Salakhutdinov, I.; Omolikov, A.; Utaganov, S. Traditional and current-prospective methods of agricultural plant diseases detection: A review. In Proceedings of the IOP Conference series: earth and environmental science, 2022; p. 012002.
- Dayarathne, M.C.; Mridha, A.U.; Wang, Y. Diagnosis of fungal plant pathogens using conventional and molecular approaches. In Diagnostics of plant diseases; IntechOpen: 2020.
- Da Lio, D.; Cobo-Díaz, J.F.; Masson, C.; Chalopin, M.; Kebe, D.; Giraud, M.; Verhaeghe, A.; Nodet, P.; Sarrocco, S.; Le Floch, G. Combined metabarcoding and multi-locus approach for genetic characterization of Colletotrichum species associated with common walnut (Juglans regia) anthracnose in France. Scientific reports 2018, 8, 10765. [Google Scholar] [CrossRef]
- Bhunjun, C.S.; Phukhamsakda, C.; Jayawardena, R.S.; Jeewon, R.; Promputtha, I.; Hyde, K.D. Investigating species boundaries in Colletotrichum. Fungal Diversity 2021, 107, 107–127. [Google Scholar] [CrossRef]
- Fuentes-Aragón, D.; Guarnaccia, V.; Rebollar-Alviter, A.; Juárez-Vázquez, S.B.; Aguirre-Rayo, F.; Silva-Rojas, H.V. Multilocus identification and thiophanate-methyl sensitivity of Colletotrichum gloeosporioides species complex associated with fruit with symptoms and symptomless leaves of mango. Plant Pathology 2020, 69, 1125–1138. [Google Scholar] [CrossRef]
- Du JunBo, D.J.; Han TianFu, H.T.; Gai JunYi, G.J.; Yong TaiWen, Y.T.; Sun Xin, S.X.; Wang XiaoChun, W.X.; Yang Feng, Y.F.; Liu Jiang, L.J.; Shu Kai, S.K.; Liu WeiGuo, L.W. Maize-soybean strip intercropping: achieved a balance between high productivity and sustainability. 2018.
- Yang, F.; Wang, X.; Liao, D.; Lu, F.; Gao, R.; Liu, W.; Yong, T.; Wu, X.; Du, J.; Liu, J. Yield response to different planting geometries in maize–soybean relay strip intercropping systems. Agronomy Journal 2015, 107, 296–304. [Google Scholar] [CrossRef]
- Liu, J.; Deng, J.; Zhang, K.; Wu, H.; Yang, C.; Zhang, X.; Du, J.; Shu, K.; Yang, W. Pod mildew on soybeans can mitigate the damage to the seed arising from field mold at harvest time. Journal of agricultural and food chemistry 2016, 64, 9135–9142. [Google Scholar] [CrossRef] [PubMed]
- Doohan, F.; Brennan, J.; Cooke, B. Influence of climatic factors on Fusarium species pathogenic to cereals. Epidemiology of Mycotoxin Producing Fungi: Under the Aegis of COST Action 835 ‘Agriculturally Important Toxigenic Fungi 1998–2003’, EU project (QLK 1-CT-1998–01380) 2003, 755-768.
- Liu, Y.; Wu, D.; Liu, Q.; Zhang, S.; Tang, Y.; Jiang, G.; Li, S.; Ding, W. The sequevar distribution of Ralstonia solanacearum in tobacco-growing zones of China is structured by elevation. European Journal of Plant Pathology 2017, 147, 541–551. [Google Scholar] [CrossRef]
- Jiang, G.; Wei, Z.; Xu, J.; Chen, H.; Zhang, Y.; She, X.; Macho, A.P.; Ding, W.; Liao, B. Bacterial wilt in China: history, current status, and future perspectives. Frontiers in Plant Science 2017, 8, 1549. [Google Scholar] [CrossRef]
- Zhou, Y.; Gong, G.; Cui, Y.; Zhang, D.; Chang, X.; Hu, R.; Liu, N.; Sun, X. Identification of Botryosphaeriaceae species causing kiwifruit rot in Sichuan Province, China. Plant Disease 2015, 99, 699–708. [Google Scholar] [CrossRef]
- O’Donnell, K.; Humber, R.A.; Geiser, D.M.; Kang, S.; Park, B.; Robert, V.A.; Crous, P.W.; Johnston, P.R.; Aoki, T.; Rooney, A.P. Phylogenetic diversity of insecticolous fusaria inferred from multilocus DNA sequence data and their molecular identification via FUSARIUM-ID and Fusarium MLST. Mycologia 2012, 104, 427–445. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular biology and evolution 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Gao, X.; Wu, M.; Xu, R.; Wang, X.; Pan, R.; Kim, H.-J.; Liao, H. Root interactions in a maize/soybean intercropping system control soybean soil-borne disease, red crown rot. Plos one 2014, 9, e95031. [Google Scholar] [CrossRef]
- Naeem, M.; Munir, M.; Li, H.; Raza, M.A.; Song, C.; Wu, X.; Irshad, G.; Khalid, M.H.B.; Yang, W.; Chang, X. Transcriptional responses of Fusarium graminearum interacted with soybean to cause root rot. Journal of Fungi 2021, 7, 422. [Google Scholar] [CrossRef]
- Miedaner, T.; Bolduan, C.; Melchinger, A. Aggressiveness and mycotoxin production of eight isolates each of Fusarium graminearum and Fusarium verticillioides for ear rot on susceptible and resistant early maize inbred lines. European Journal of Plant Pathology 2010, 127, 113–123. [Google Scholar] [CrossRef]
- Chang, X.; Wei, D.; Zeng, Y.; Zhao, X.; Hu, Y.; Wu, X.; Song, C.; Gong, G.; Chen, H.; Yang, C. Maize-soybean relay strip intercropping reshapes the rhizosphere bacterial community and recruits beneficial bacteria to suppress Fusarium root rot of soybean. Frontiers in Microbiology 2022, 13, 1009689. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.J.; Chen, H.K.; Ni, H.-F. Identification and characterization of Colletotrichum species associated with mango anthracnose in Taiwan. European Journal of Plant Pathology 2020, 157, 1–15. [Google Scholar] [CrossRef]
- Cacciola, S.O.; Gilardi, G.; Faedda, R.; Schena, L.; Pane, A.; Garibaldi, A.; Gullino, M.L. Characterization of Colletotrichum ocimi population associated with black spot of sweet basil (Ocimum basilicum) in Northern Italy. Plants 2020, 9, 654. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Gilardi, G.; Martino, I.; Garibaldi, A.; Gullino, M.L. Species diversity in Colletotrichum causing anthracnose of aromatic and ornamental Lamiaceae in Italy. Agronomy 2019, 9, 613. [Google Scholar] [CrossRef]
- Liu, F.; Damm, U.; Cai, L.; Crous, P.W. Species of the Colletotrichum gloeosporioides complex associated with anthracnose diseases of Proteaceae. Fungal Diversity 2013, 61, 89–105. [Google Scholar] [CrossRef]
- Summerell, B.A. Resolving Fusarium: Current status of the genus. Annual review of phytopathology 2019, 57, 323–339. [Google Scholar] [CrossRef]
- O’Donnell, K.; Whitaker, B.K.; Laraba, I.; Proctor, R.H.; Brown, D.W.; Broders, K.; Kim, H.S.; McCormick, S.P.; Busman, M.; Aoki, T. DNA sequence-based identification of Fusarium: A work in progress. Plant disease 2022, 106, 1597–1609. [Google Scholar] [CrossRef]
- Neergaard, P.; Neergaard, P. Management of Seed Storage. Seed Pathology: Volume I 1977, 574-594.
- Killebrew, J.; Roy, K.; Lawrence, G.; McLean, K.; Hodges, H. Greenhouse and field evaluation of Fusarium solani pathogenicity to soybean seedlings. 1988.
- Feng, J.; Hwang, R.; Chang, K.; Hwang, S.; Strelkov, S.; Gossen, B.; Conner, R.; Turnbull, G. Genetic variation in Fusarium avenaceum causing root rot on field pea. Plant Pathology 2010, 59, 845–852. [Google Scholar] [CrossRef]
- van Diepeningen, A.D.; Brankovics, B.; Iltes, J.; Van der Lee, T.A.; Waalwijk, C. Diagnosis of Fusarium infections: approaches to identification by the clinical mycology laboratory. Current fungal infection reports 2015, 9, 135–143. [Google Scholar] [CrossRef]
| Fusarium species | Macroconidia | Colony characterization | Growth rate (cm/day) | |||
|---|---|---|---|---|---|---|
| Shape | Width (μm) | Length (μm) | Septum | |||
| F. equiseti | Falcate | 3.10±0.02c, 3.02-3.7 |
39.25±1.81a, 38.23-45.98 |
3-5 | Pale grey color (front), ginger yellowish (back) | 4.88 ± 0.41b |
| F. incarnatum | Falcate | 3.98±0.44a, 5.67-2.72 |
36.98±3.63a, 45.55-36.62 |
3-4 | Pale grey color(front), yellowish color (back) | 5.32 ± 0.39a |
| F. verticillioides | Fusiform | 3.40±0.9b, 3.8-3.33 |
23.25±0.2b, 20.21-25.90 |
2-3 | pale grey colonies, reverse pale grey | 4.9 ± 0.3b |
| F. proliferatum | Falcate, fusiform | 3.60±1.12b, 5.41-2.96 |
39.12±6.54a, 48.56-32.66 |
3-4 | Pale grey color (front), pale grey (back) | 4.50 ± 0.03c |
| F. fujikuroi | Falcate | 2.42±0.46e, 3.12-2.28 |
39.92±1.98a, 43.82-38.94 |
3.-5 | Pale grey color (front), pale yellowish color (back) | 4.76 ± 0.32c |
| F. oxysporum | Falcate, with a foot spore | 3.1±0.16c, 2.31-4.82 |
26.9±1.6b, 28.62-22.23 |
3 | pale grey (front) pale purple on the back | 5.4 ± 0.3a |
| F. chlamydosporum | Falcate | 3.20±0.82c, 3.90-3.22 |
25.45±0.20b, 28.12-23.95 |
2-3 | Brown, light pink (Front) purple (back) | 5.23 ± 0.01a |
| F. acutatum | Falcate | 2.79±1.62d, 3.20-1.98 |
23.24±0.8b, 24.56-20.86 |
3-5 | white grey (front) purple (back) |
4.39 ± 0.02c |
| Species name | Texture | Conidial shape | Conidia size | Growth rate (cm/day) |
|
|---|---|---|---|---|---|
| Length (μm) | Width (μm) | ||||
| C. truncatum | Cottony | Fusiform | 23.20±0.56a 24.65-16.22 |
5.56±0.35b 5.90-4.32 |
6.90 ± 0.12a |
| C. karstii | Cottony | Cylindrical | 15.5±0.20b 18.20-14.90 |
6.80±0.23a 8.56-5.52 |
5.63 ± 0.27b |
| C. cliviicola | Cottony | oval/ellipsoidal | 13.35±0.02c 14.22-12.86 |
3.62±0.06c 4.56-3.22 |
6.12 ± 0.25a |
| C. plurivorum | Cottony and white | Fusiform | 13.75±0.12c 15.78-12.66 |
3.4±0.02c 4.45-3.56 |
6.30 ± 0.09a |
| C. boninense | Medium brown | Cylindrical | 15.10±0.2b 16.20-14.75 |
5.30±0.45b 6.60-4.25 |
6.15±0.02a |
| C.fructicola | Greyish black | Fusiform | 12.90±0.32c 14.56-10.86 |
6.80±0.23a 8.56-5.52 |
6.4±0.60a |
| Isolates | Disease incidence (%) | PMC (%) | DSI (%) | Pod Weight (g) |
|---|---|---|---|---|
| Control (CK) | 0 | 0±0e | 0±0d | 1.89±0.04d |
| F. proliferatum (FS31) | 100 | 90.66±0.40a | 96.66±4.71a | 1.79±0.31c |
| F. proliferatum (FS120) | 100 | 89.66±0.47a | 100±0a | 1.71±0.05c |
| F. proliferatum (FS167) | 100 | 78.33±2.35a | 93.33±4.71a | 1.74±0.01c |
| F. fujikuroi (FS79) | 100 | 43.33±2.35b | 66.66±11.78b | 2.48±0.27b |
| F. fujikuroi (FS101) | 100 | 46.66±4.71b | 75±0ab | 2.37±0.24b |
| F. fujikuroi (FS123) | 100 | 23.33±2.35c | 58.33±11.78b | 2.71±0.09b |
| F. equiseti (FS4) | 100 | 88.33±2.35a | 75±11.78b | 2.21±0.07b |
| F. equiseti (FS65) | 100 | 41.66±2.35b | 50±11.78b | 2.19±0.02b |
| F. equiseti (FS170) | 100 | 21.66±2.35c | 50±20.41b | 2.13±0.03b |
| F. acutatum (FS142) | 100 | 100±0a | 100±0a | 2.64±0.09b |
| F. acutatum (FS151) | 100 | 95±4.08a | 100±0a | 2.47±0.31b |
| F. acutatum (FS155) | 100 | 98.33±2.35a | 100±0a | 2.57±0.12b |
| F. verticillioides (FS42) | 100 | 98.33±2.35a | 100±0a | 2.29±0.12b |
| F. verticillioides (FS89) | 100 | 100±0a | 100±0a | 2.25±0.04b |
| F. verticillioides (FS112) | 100 | 100±0a | 100±0a | 2.29±0.06c |
| F. incarnatum (FS2) | 100 | 26.66±2.35c | 33.33±11.78c | 3.34±0.19a |
| F. incarnatum (FS24) | 100 | 20±4.08dc | 25±0c | 3.29±0.05a |
| F. incarnatum (FS130) | 100 | 18.33±6.23d | 33.33±11.78c | 3.25±0.06a |
| F. oxysporum (FS49) | 100 | 55±4.08b | 83.33±11.78a | 1.84±0.27c |
| F. oxysporum (FS26) | 100 | 45±4.08b | 66.66±11.78b | 1.86±0.04c |
| F. oxysporum (FS132) | 100 | 50±4.08b | 58.33±11.78b | 1.89±0.16c |
| F. chlamydosporum (FS76) | 100 | 18.33±2.35d | 75±0ab | 1.68±0.16c |
| F. chlamydosporum (FS105) | 100 | 23.33±6.23c | 50±20.41b | 1.57±0.05c |
| F. chlamydosporum (FS126) | 100 | 16.66±2.35d | 66.66±11.78b | 1.69±0.08c |
| Isolates | Disease incidence (%) | PMC (%) | DSI (%) | Pod weight (g) |
|---|---|---|---|---|
| Control (CK) | 0 | 0±0d | 0±0e | 1.86±0.040d |
| C. fructicola (CS15) | 100 | 90±4.08a | 100±0a | 2.51±0.04b |
| C. fructicola (CS93) | 100 | 86.66±6.23a | 91.66±11.78a | 2.52±0.02b |
| C. fructicola (CS169) | 100 | 76.66±2.35ab | 100±0a | 2.51±0.01b |
| C. truncatum (CS03) | 100 | 78.33±4.71ab | 75±0b | 2.09±0.02c |
| C. truncatum (CS61) | 100 | 80±4.08a | 83.33±11.78b | 2.11±0.01c |
| C. truncatum (CS153) | 100 | 81.66±2.35a | 66.66±11.7b | 2.16±0.08c |
| C. karstii (CS13) | 100 | 91.66±2.35a | 83.33±11.78b | 2.67±0.05b |
| C. karstii (CS 96) | 100 | 86.66±2.35a | 75±0b | 2.47±0.08b |
| C. karstii (CS 158) | 100 | 80±7.07a | 75±0b | 2.67±0.03b |
| C. cliviicola (CS 22) | 100 | 81.66±2.35a | 66.66±11.78c | 2.13±0.12c |
| C. cliviicola (CS95) | 100 | 76.66±2.35ab | 58.33±11.78c | 2.21±0.08c |
| C. cliviicola (CS 164) | 100 | 65±4.0b | 50±20.41c | 2.26±0.09c |
| C. plurivorum (CS06) | 100 | 10±4.08c | 25±0d | 2.60±0.02b |
| C. plurivorum (CS172) | 100 | 8.33±2.35c | 33.33±11.78d | 2.59±0.01b |
| C. plurivorum (CS180) | 100 | 6.66±2.35c | 25±20.41d | 2.61±0.01b |
| C. boninense (CS94) | 100 | 10±4.08c | 33.33±11.78d | 2.96±0.01a |
| C. boninense (CS118) | 100 | 11.66±6.23c | 25±0d | 2.98±0.02a |
| C. boninense (CS166) | 100 | 10±4.08c | 25±0d | 2.95±0.03a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





