Submitted:
23 July 2025
Posted:
24 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Patient Cohort and Study Design
2.2. Patient Selection
2.3. DNA Extraction
2.4. BRCA Testing with Myriapod® NGS BRCA1/2 Panel Kit and Primary Sequencing Strategy
2.5. Re-Evaluation of CNV Calling Using Diatech Software by Simulating a Diagnostic Setting
2.6. Read Coverage and Comparative Analyses
2.7. Data Analysis
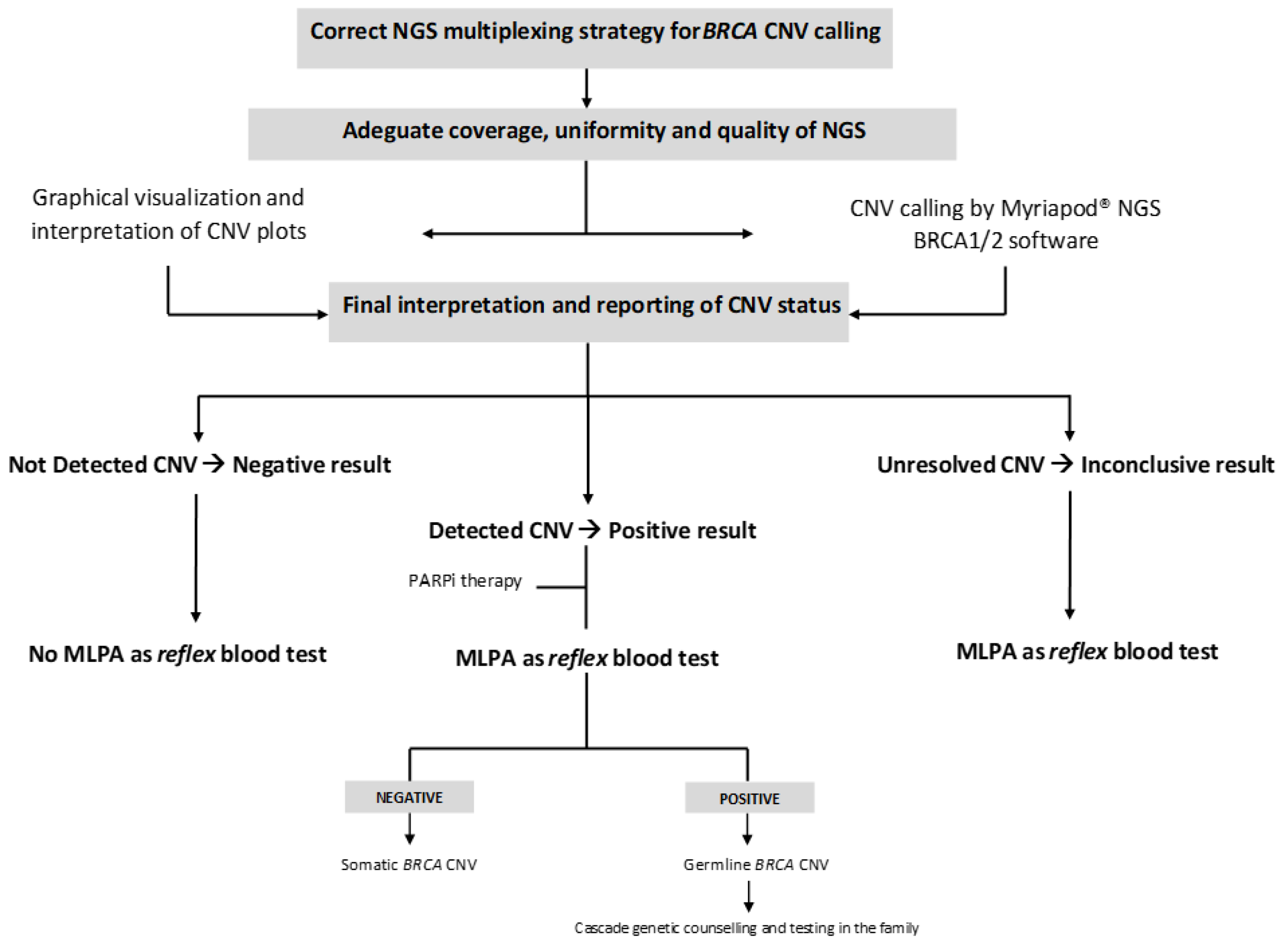
3. Results
- (a)
- Graphical visualization and interpretation of CNV plots;
- (b)
- CNV calling by the Myriapod® NGS data analysis software;
- (c)
- Final interpretation and reporting of CNV status, as a decision-making result integrating the two previous analysis levels.
3.1. Concordance Analysis Between TSO500HT/SOPHiA DDM HRD and Myriapod® NGS BRCA 1-2 Pipeline in Primary CNV Calling Strategy (5/4 BRCA CNV-Positive vs 5/6 BRCA CNV-Negative Samples in the Same NGS Run)
3.1.1. BRCA-CNV-Negative Samples
3.1.2. BRCA-CNV-Positive Samples
3.2. CNV Calling in a Simulated Diagnostic Scenario (1 BRCA CNV-Positive vs 9 BRCA CNV-Negative Samples in the Same NGS Run)
3.2.1. BRCA-CNV-Negative Samples
3.2.2. BRCA-CNV Positive Samples
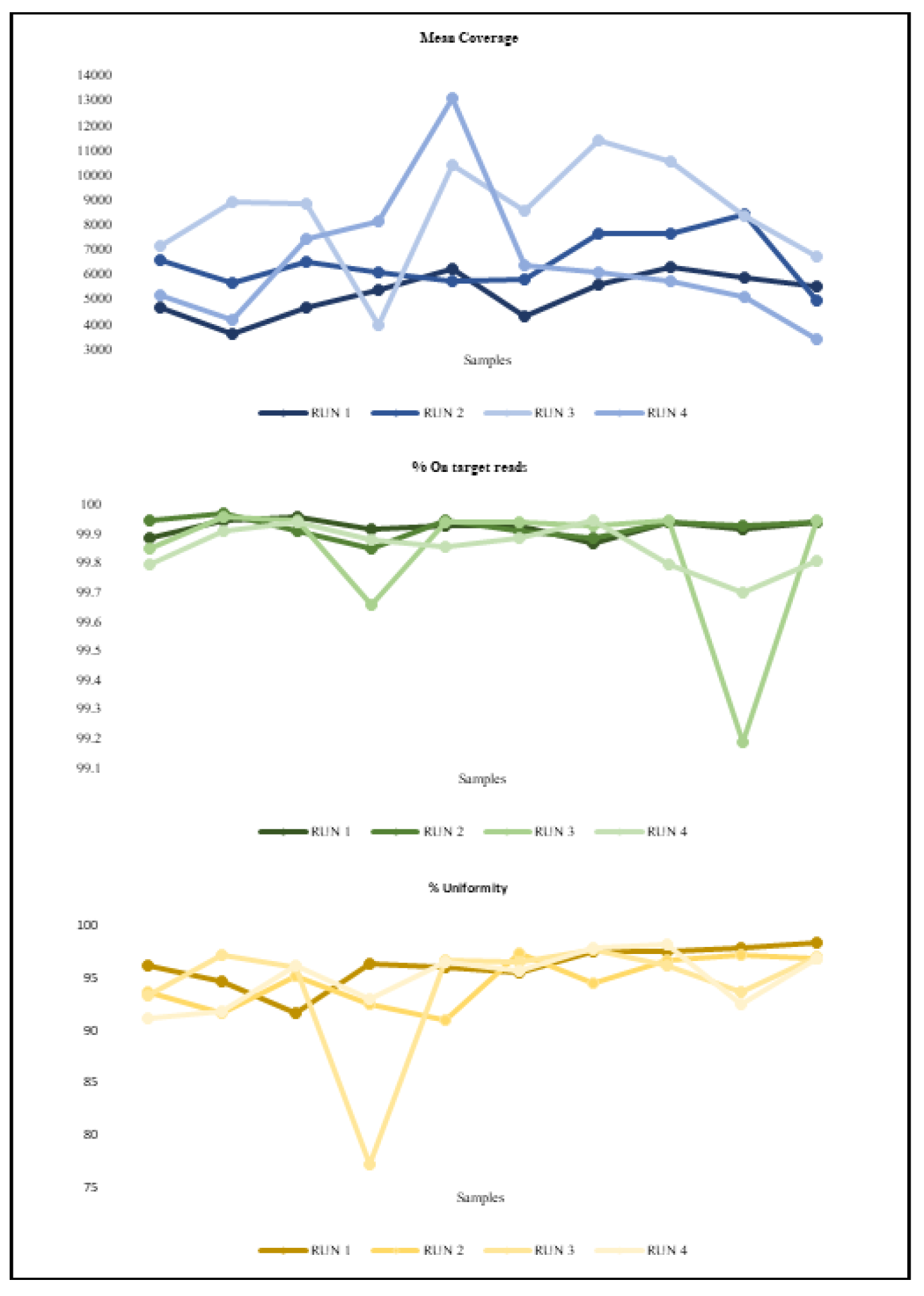
3.3. Sequencing Metrics and Performance
4. Discussion
5. Conclusions
Acknowledgements
Conflicts of Interest
Abbreviations
References
- Marmolejo, D.H.; Wong, M.Y.Z. Overview of Hereditary Breast and Ovarian Cancer (HBOC) Guidelines across Europe. Eur. J. Med. Genet. 2021, 64, 104350.
- Lee, J.M.; Ledermann, J.A.; Kohn, E.C. PARP Inhibitors for BRCA1/2 Mutation-Associated and BRCA-Like Malignancies. Ann. Oncol. 2014, 25, 32–40.
- Dewani, D.; Jaiswal, A.; Karwade, P.C. Poly(Adenosine Diphosphate Ribose) Polymerase (PARP) Inhibitors in the Treatment of Advanced Ovarian Cancer: A Narrative Review. Cureus 2024, 16, e68463.
- Guffanti, F.; Mengoli, I.; Damia, G. Current HRD Assays in Ovarian Cancer: Differences, Pitfalls, Limitations, and Novel Approaches. Front. Oncol. 2024, 14, 1405361.
- Ratnaparkhi, R.; Javellana, M.; Jewell, A.; Spoozak, L. Evaluation of Homologous Recombination Deficiency in Ovarian Cancer. Curr. Treat. Options Oncol. 2024, 25, 237–260.
- Arcieri, M.; Tius, V.; Andreetta, C.; Restaino, S.; Biasioli, A.; Poletto, E.; Damante, G.; Ercoli, A.; Driul, L.; Fagotti, A.; Lorusso, D.; Scambia, G.; Vizzielli, G. How BRCA and Homologous Recombination Deficiency Change Therapeutic Strategies in Ovarian Cancer: A Review of Literature. Front. Oncol. 2024, 14, 1335196.
- Mafficini, A.; Simbolo, M.; Parisi, A.; Rusev, B.; Luchini, C.; Cataldo, I.; Piazzola, E.; Sperandio, N.; Turri, G.; Franchi, M.; Tortora, G.; Bovo, C.; Lawlor, R.T.; Scarpa, A. BRCA Somatic and Germline Mutation Detection in Paraffin-Embedded Ovarian Cancers by Next-Generation Sequencing. Oncotarget 2016, 7, 1076–1083.
- Ewald, I.P.; Ribeiro, P.L.; Palmero, E.I.; Cossio, S.L.; Giugliani, R.; Ashton-Prolla, P. Genomic Rearrangements in BRCA1 and BRCA2: A Literature Review. Genet. Mol. Biol. 2009, 32, 437–446.
- Concolino, P.; Mello, E.; Minucci, A.; Santonocito, C.; Scambia, G.; Giardina, B.; Capoluongo, E. Advanced Tools for BRCA1/2 Mutational Screening: Comparison between Two Methods for Large Genomic Rearrangements (LGRs) Detection. Clin. Chem. Lab. Med. 2014, 52, 1119–1127.
- Enyedi, J.; Pintér, L.; Sükösd, F.; Gyuris, Z.; Hajdu, A.; Határvölgyi, E.; Priskin, K.; Haracska, L. Simultaneous Detection of BRCA Mutations and Large Genomic Rearrangements in Germline DNA and FFPE Tumor Samples. Oncotarget 2016, 20, 61845–61859.
- Schmidt, A.Y.; Hansen, T.V.O.; Ahlborn, L.B.; Jønson, L.; Yde, C.W.; Nielsen, F.C. Next-Generation Sequencing-Based Detection of Germline Copy Number Variations in BRCA1/BRCA2: Validation of a One-Step Diagnostic Workflow. J. Mol. Diagn. 2017, 19, 809–816.
- Minucci, A.; Mazzuccato, G.; Marchetti, C.; Pietragalla, A.; Scambia, G.; Fagotti, A.; Urbani, A. Detecting Large Germline Rearrangements of BRCA1 by Next-Generation Tumor Sequencing. Mol. Biol. (Mosk.) 2020, 54, 688–698.
- Nero C.; Duranti S.; Giacomini F.; Minucci A.; Giacò L.; Piermattei A.; Genuardi M.;Pasciuto T.; Urbani A.; Daniele G.; Lorusso D.; Pignataro R.; Tortora G.; Normanno N.; Scambia G. Integrating a Comprehensive Cancer Genome Profiling into Clinical Practice: A Blueprint in an Italian Referral Center. J. Pers. Med. 2022,12,1746.
- Minucci, A.; Scambia, G.; Santonocito, C.; Concolino, P.; Canu, G.; Mignone, F.; Saggese, I.; Guarino, D.; Costella, A.; Molinario, R.; De Bonis, M.; Ferrandina, G.; Petrillo, M.; Scaglione, G.L.; Capoluongo, E. Clinical Impact on Ovarian Cancer Patients of Massive Parallel Sequencing for BRCA Mutation Detection: The Experience at Gemelli Hospital and a Literature Review. Expert Rev. Mol. Diagn. 2015, 15, 1383–1403.
- Szczerba, E.; Kamińska, K.; Mierzwa, T.; Misiek, M.; Kowalewski, J.; Lewandowska, M.A. BRCA1/2 Mutation Detection in the Tumor Tissue from Selected Polish Patients with Breast Cancer Using Next Generation Sequencing. Genes 2021, 12, 519.
- Barili, V.; Ambrosini, E.; Bortesi, B.; Minari, R.; De Sensi, E.; Cannizzaro, I.R.; Taiani, A.; Michiara, M.; Sikokis, A.; Boggiani, D.; et al. Genetic Basis of Breast and Ovarian Cancer: Approaches and Lessons Learnt from Three Decades of Inherited Predisposition Testing. Genes 2024, 15, 219.
| ID | Disease | Timing | Age of the sample | Tumor content (%) | CNV Status | Reference assay | Gene | Exons | Type of CNV | Status of CNV* |
| 1 | HGSC | PRIMARY | 2024 | 90 | Positive | TSO500HT | BRCA2 | 2-3 | Intragenic deletion | Somatic |
| 2 | HGSC | PRIMARY | 2024 | 90 | Positive | TSO500HT SOPHiA DDM HRD HRD |
BRCA1 | 19 | Intragenic deletion | Somatic |
| 3 | HGSC | PRIMARY | 2024 | 60 | Positive | TSO500HT SOPHiA DDM HRD |
BRCA1 | 8-11 | Intragenic deletion | Germline |
| 4 | HGSC | PRIMARY | 2024 | 70 | Positive | TSO500HT SOPHiA DDM HRD |
BRCA1 | 20 | Intragenic deletion | Germline |
| 5 | HGSC | RELAPSE | 2023 | 90 | Positive | TSO500HT SOPHiA DDM HRD |
BRCA1 | 2 | Intragenic deletion | Germline |
| 6 | HGSC | RELAPSE | 2024 | 90 | Positive | SOPHiA DDM HRD | BRCA1 | 16-17 | Intragenic deletion | Germline |
| 7 | HGSC | RELAPSE | 2024 | 70 | Positive | TSO500HT SOPHiA DDM HRD |
BRCA1 | 4-7 | Intragenic deletion | Germline |
| 8 | HGSC | PRIMARY | 2023 | 90 | Positive | TSO500HT SOPHiA DDM HRD |
BRCA2 | 19-21 | Intragenic deletion | Somatic |
| 9 | ENOC | PRIMARY | 2024 | 90 | Positive | TSO500HT | BRCA1 | 11 | Intragenic deletion | Somatic |
| 10 | HGSC | PRIMARY | 2024 | 95 | Positive | TSO500HT SOPHiA DDM HRD |
BRCA1 | 2-3 | Intragenic deletion | Somatic |
| 11 | HGSC | PRIMARY | 2024 | 60 | Positive | TSO500HT SOPHiA DDM HRD |
BRCA1 | 19 | Intragenic deletion | Germline |
| 12 | HGSC | RELAPSE | 2024 | 80 | Positive | SOPHiA DDM HRD | BRCA1 | 2 | Intragenic deletion | Germline |
| 13 | HGSC | RELAPSE | 2024 | 80 | Positive | SOPHiA DDM HRD | BRCA1 | 2- 19 | Intragenic deletion | Somatic |
| 14 | HGSC | PRIMARY | 2024 | 80 | Positive | TSO500HT | BRCA1 | 15 | Intragenic deletion | Somatic |
| 15 | HGSC | PRIMARY | 2024 | 55 | Positive | TSO500HT SOPHiA DDM HRD |
BRCA1 | 1-24 | Whole gene deletion | Germline |
| 16 | HGSC | PRIMARY | 2025 | 30 | Positive | TSO500HT SOPHiA DDM HRD |
BRCA2 | 11-27 | Intragenic deletion | Somatic |
| 17 | HGSC | PRIMARY | 2024 | 80 | Positive | TSO500HT | BRCA1 | 3-23 | Intragenic deletion | Somatic |
| 18 | OCS | RELAPSE | 2024 | 60 | Positive | TSO500HT SOPHiA DDM HRD |
- | - | - | - |
| 19 | HGSC | PRIMARY | 2024 | 80 | Negative | TSO500HT SOPHiA DDM HRD |
- | - | - | - |
| 20 | HGSC | PRIMARY | 2025 | 20 | Negative | TSO500HT | - | - | ||
| 21 | HGSC | PRIMARY | 2024 | 80 | Negative | TSO500HT SOPHiA DDM HRD |
- | - | - | - |
| 22 | HGSC | PRIMARY | 2024 | 80 | Negative | TSO500HT | - | - | - | - |
| 23 | CCOC | PRIMARY | 2024 | 80 | Negative | TSO500HT | - | - | - | - |
| 24 | ENOC | PRIMARY | 2024 | 90 | Negative | TSO500HT | - | - | - | - |
| 25 | HGSC | PRIMARY | 2024 | 25 | Negative | TSO500HT | - | - | - | - |
| 26 | HGSC | PRIMARY | 2024 | 80 | Negative | TSO500HT SOPHiA DDM HRD |
- | - | - | - |
| 27 | HSGC | RELAPSE | 2024 | 95 | Negative | SOPHiA DDM HRD | - | - | - | - |
| 28 | ENOC | RELAPSE | 2024 | 30 | Negative | SOPHiA DDM HRD | - | - | - | - |
| 29 | HSGC | RELAPSE | 2024 | 30 | Negative | SOPHiA DDM HRD | - | - | - | - |
| 30 | HSGC | RELAPSE | 2024 | 35 | Negative | SOPHiA DDM HRD | - | - | - | - |
| 31 | HGSC | PRIMARY | 2023 | 70 | Negative | TSO500HT SOPHiA DDM HRD |
- | - | - | - |
| 32 | CCOC | PRIMARY | 2023 | 80 | Negative | TSO500HT | - | - | - | - |
| 33 | HGSC | PRIMARY | 2024 | 40 | Negative | TSO500HT SOPHiA DDM HRD |
- | - | - | - |
| 34 | HGSC | PRIMARY | 2024 | 36 | Negative | SOPHiA DDM HRD | - | - | - | - |
| 35 | HGSC | PRIMARY | 2023 | 80 | Negative | SOPHiA DDM HRD | - | - | - | - |
| 36 | HGSC | PRIMARY | 2025 | 70 | Negative | TSO500HT SOPHiA DDM HRD |
- | - | - | - |
| 37 | CCOC | PRIMARY | 2025 | 20 | Negative | TSO500HT | - | - | - | - |
| 38 | HGSC | PRIMARY | 2025 | 30 | Negative | TSO500HT SOPHiA DDM HRD |
- | - | - | - |
| 39 | CCOC | PRIMARY | 2025 | 70 | Negative | TSO500HT | - | - | - | - |
| 40 | HGSC | PRIMARY | 2025 | 25 | Negative | TSO500HT | - | - | - | - |
|
ID samples |
Graphical visualization and intepretation of CNV plots |
CNV calling by Diatech software |
Final interpretation and reporting CNV |
||||||||||
| BRCA1 | BRCA2 | BRCA1 | BRCA2 | BRCA1 | BRCA1 | ||||||||
| BRCA CNV-negative samples | |||||||||||||
| 18 | cCNV | oCNV | Potential CNV | Potential CNV | Negative | oCNV | |||||||
| 19 | oCNV | cCNV | Potential CNV | CNV Not Positive | Negative | Negative | |||||||
| 20 | cCNV | cCNV | CNV Not Positive | Potential CNV | Negative | Negative | |||||||
| 21 | cCNV | cCNV | Potential CNV | Potential CNV | Negative | Negative | |||||||
| 22 | cCNV | cCNV | Potential CNV | CNV Not Positive | Negative | Negative | |||||||
| 23 | cCNV | cCNV | Potential CNV | CNV Not Positive | Negative | Negative | |||||||
| 24 | cCNV | cCNV | Potential CNV | CNV Not Positive | Negative | Negative | |||||||
| 25 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | |||||||
| 26 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | |||||||
| 27 | oCNV | cCNV | Potential CNV | CNV Not Positive | oCNV | Negative | |||||||
| 28 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | |||||||
| 29 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | |||||||
| 30 | oCNV | cCNV | Potential CNV | CNV Not Positive | oCNV | Negative | |||||||
| 31 | cCNV | cCNV | Potential CNV | CNV Not Positive | Negative | Negative | |||||||
| 32 | cCNV | cCNV | Potential CNV | CNV Not Positive | Negative | Negative | |||||||
| 33 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | |||||||
| 34 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | |||||||
| 35 | cCNV | cCNV | CNV Not Positive | Potential CNV | Negative | Negative | |||||||
| 36 | cCNV | cCNV | CNV Not Positive | Potential CNV | Negative | Negative | |||||||
| 37 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | |||||||
| 38 | iCNV | cCNV | Potential CNV | CNV Not Positive | Negative | Negative | |||||||
| 39 | cCNV | iCNV | CNV Not Positive | Potential CNV | Negative | iCNV | |||||||
| 40 | oCNV | cCNV | Potential CNV | Potential CNV | oCNV | Negative | |||||||
| BRCA CNV-positive samples | |||||||||||||
| 1 | oCNV | cCNV | Potential CNV | Potential CNV | Positive | Positive | |||||||
| 2 | cCNV | cCNV | Potential CNV | Potential CNV | Positive | Negative | |||||||
| 3 | cCNV | cCNV | Potential CNV | Potential CNV | Positive | Negative | |||||||
| 4 | cCNV | cCNV | Potential CNV | CNV Not Positive | Positive | Negative | |||||||
| 5 | cCNV | cCNV | Potential CNV | CNV Not Positive | Positive | Negative | |||||||
| 6 | cCNV | cCNV | CNV Not Positive | Potential CNV | Positive | Negative | |||||||
| 7 | cCNV | oCNV | CNV Not Positive | Potential CNV | Negative | iCNV | |||||||
| 8 | oCNV | cCNV | Potential CNV | Potential CNV | oCNV | Positive | |||||||
| 9 | fCNV | fCNV | CNV Failed | CNV Failed | fCNV | fCNV | |||||||
| 10 | cCNV | cCNV | CNV Not Positive | Potential CNV | Positive | Negative | |||||||
| 11 | cCNV | cCNV | Potential CNV | CNV Not Positive | Positive | Negative | |||||||
| 12 | cCNV | iCNV | CNV Not Positive | Potential CNV | Positive | iCNV | |||||||
| 13 | cCNV | cCNV | CNV Not Positive | Potential CNV | Positive | Negative | |||||||
| 14 | cCNV | cCNV | Potential CNV | Potential CNV | Positive | Positive | |||||||
| 15 | ntCNV | iCNV | CNV Not Positive | Potential CNV | Negative | iCNV | |||||||
| 16 | cCNV | cCNV | CNV Not Positive | Potential CNV | Negative | Positive | |||||||
| 17 | ntCNV | iCNV | CNV Not Positive | Potential CNV | iCNV | iCNV | |||||||
| A) Primary CNV Calling Strategy and concordance analysis (5/4 BRCA CNV-positive vs 5/6 BRCA CNV-negative samples). | ||
| Diatech Myriapod® NGS BRCA1/2panel kit | TSO500HT/SOPHiA DDM™ HRD | |
| CNV Positive | CNV Negative | |
| CNV Positive | 13 | 5 |
| CNV Negative | 3 | 18 |
| Inconclusive | 1 | |
| Analytical Performance | Value (%) | |
| Sensitivity | 81.25 | |
| Specificity | 78.26 | |
| Positive predictive value | 72.22 | |
| Negative predictive value | 85.71 | |
| Accuracy | 79.49 | |
| B) CNV Calling in a simulated BRCA diagnostic setting (1 BRCA CNV-positive vs 9 BRCA CNV-negative samples) | ||
| Diatech Myriapod® NGS BRCA1/2panel kit | TSO500HT/SOPHiA DDM™ HRD | |
| CNV Positive | CNV Negative | |
| CNV Positive | 15 | 1 |
| CNV Negative | 1 | 22 |
| Inconclusive | 1 | |
| Analytical Performance | Value (%) | |
| Sensitivity | 93.75 | |
| Specificity | 95.65 | |
| Positive predictive value | 94.87 | |
| Negative predictive value | 93.75 | |
| Accuracy | 95.65 | |
|
ID samples |
Graphical visualization and intepretation of CNV plots |
CNV calling by Diatech software |
Final interpretation and reporting CNV |
|||||||
| BRCA1 | BRCA2 | BRCA1 | BRCA2 | BRCA1 | BRCA1 | |||||
| BRCA CNV-negative samples | ||||||||||
| 18 | cCNV | cCNV | CNV Not Positive | Potential CNV | Negative | Negative | ||||
| 19 | cCNV | cCNV | CNV Not Positive | Potential CNV | Negative | Negative | ||||
| 20 | cCNV | cCNV | Potential CNV | CNV Not Positive | Negative | Negative | ||||
| 21 | cCNV | cCNV | CNV Not Positive | Potential CNV | Negative | Negative | ||||
| 22 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | ||||
| 23 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | ||||
| 24 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | ||||
| 25 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | ||||
| 26 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | ||||
| 27 | cCNV | cCNV | Potential CNV | CNV Not Positive | Negative | Negative | ||||
| 28 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | ||||
| 29 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | ||||
| 30 | cCNV | cCNV | Potential CNV | CNV Not Positive | Negative | Negative | ||||
| 31 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | ||||
| 32 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | ||||
| 33 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | ||||
| 34 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | ||||
| 35 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | ||||
| 36 | cCNV | cCNV | CNV Not Positive | Potential CNV | Negative | Negative | ||||
| 37 | cCNV | cCNV | CNV Not Positive | CNV Not Positive | Negative | Negative | ||||
| 38 | cCNV | cCNV | Potential CNV | CNV Not Positive | Negative | Negative | ||||
| 39 | oCNV | cCNV | Potential CNV | Potential CNV | oCNV | Negative | ||||
| 40 | cCNV | cCNV | Potential CNV | Potential CNV | Negative | Negative | ||||
| BRCA CNV-positive samples | ||||||||||
| 1 | cCNV | cCNV | CNV Not Positive | Potential CNV | Negative | Positive | ||||
| 2 | cCNV | cCNV | Potential CNV | CNV Not Positive | Positive | Negative | ||||
| 3 | cCNV | cCNV | Potential CNV | Potential CNV | Positive | Negative | ||||
| 4 | cCNV | cCNV | Potential CNV | CNV Not Positive | Positive | Negative | ||||
| 5 | cCNV | cCNV | Potential CNV | CNV Not Positive | Positive | Negative | ||||
| 6 | cCNV | cCNV | Potential CNV | CNV Not Positive | Positive | Negative | ||||
| 7 | cCNV | iCNV | CNV Not Positive | Potential CNV | Positive | iCNV | ||||
| 8 | cCNV | cCNV | Potential CNV | Potential CNV | Negative | Positive | ||||
| 9 | fCNV | fCNV | CNV failed | CNV failed | fCNV | fCNV | ||||
| 10 | cCNV | cCNV | Potential CNV | CNV Not Positive | Positive | Negative | ||||
| 11 | cCNV | cCNV | Potential CNV | CNV Not Positive | Positive | Negative | ||||
| 12 | cCNV | cCNV | Potential CNV | Potential CNV | Positive | Negative | ||||
| 13 | cCNV | cCNV | CNV Not Positive | Potential CNV | Positive | Negative | ||||
| 14 | cCNV | cCNV | Potential CNV | Potential CNV | Positive | Negative | ||||
| 15 | cCNV | cCNV | CNV Not Positive | Potential CNV | Positive | Negative | ||||
| 16 | cCNV | cCNV | CNV Not Positive | Potential CNV | Negative | Positive | ||||
| 17 | ntCNV | iCNV | CNV Not Positive | Potential CNV | Negative | iCNV | ||||
| Mean ± Standard Deviation | |||
| ID Run | Mean Coverage | Uniformity (%) | On target reads (%) |
| 1 | 5231±876 | 96.1±1.9 | 99.9±0.03 |
| 2 | 6514±1083 | 94.6±2.3 | 99.9±0.03 |
| 3 | 8490±2167 | 94±6.12 | 99.8±0.2 |
| 4 | 6488±2732 | 94.9±2.6 | 99.8±0.07 |
| Total | 6680±1346 | 94.9±0.88 | 99.8±0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





