1. Introduction
Pre-clinical studies in the drug discovery process are conducted using animal or cell culture tests with human-derived cells to investigate a new drug candidate's efficacy, toxicity, and pharmacokinetics. However, in animal testing, there are problems of species differences between experimental animals and humans, and ethical problems [
1]
. Cell culture tests using human-derived cells have difficulties reproducing the in vivo environment and confirming organ interactions, and the extrapolation to humans is limited, making it challenging to evaluate drug efficacy accurately [
2]. Therefore, a novel drug efficacy evaluation system that can be used in pre-clinical studies is needed to replace conventional animal and cell culture tests using human-derived cells.
Recently, a novel cell culture platform called “Microphysiological System (MPS)” based on microfluidics has attracted attention [
3,
4]. MPS is an in vitro model that mimics the human in vivo environment, equipped with medium perfusion functions with microfluidic devices and a chamber capable of culturing human-derived cells [
5]. The dynamic environment provided by the perfusion in MPS can maintain the function and morphology of cultured cells and reproduce physiological functions better than conventional cell culture tests using human-derived cells [
6]. Moreover, human-derived cells used in MPS allow the evaluation of human-specific drug responses that animal testing cannot predict. MPS is designed to mimic the functions of specific organs or tissues and can evaluate the specific functions of organs or tissues in vitro. Additionally, Complex interactions between different organs are evaluated by coculturing cells from multiple organs in MPS [
7]. Based on the above, MPS is expected to be a powerful tool for evaluating new drugs' efficacy, toxicity, and pharmacokinetics. However, in drug efficacy evaluations using MPS, the medium is typically changed every 1 to 2 days to supply nutrients and remove waste products for cell maintenance. The drug concentrations in MPS change discretely because the anticancer drug in the medium is replaced simultaneously with the medium. In the in vivo environment, as the drug is absorbed, distributed, metabolized, and excreted, its concentration changes continuously over time. Reproducing continuous drug concentration changes is important for accurately predicting a drug's efficacy, toxicity, and pharmacokinetics, because the efficacy and duration of a drug depend on changes in drug concentration.
Several approaches have been proposed thus far to improve the prediction accuracy of drug efficacy and toxicity in vitro by mimicking in vivo drug concentration changes using MPS. Petreus
et al. developed a platform to accurately assess the drug efficacy of tumor spheroids by sequentially delivering eight concentrations of the drug at a constant flow rate [
8]. Additionally, Singh
et al. demonstrated that the automation of the drug injection system allows for the simulation of long-term drug concentration changes in mice [
9]. Furthermore, Guerrero
et al. used computer control to replicate in vivo drug concentration changes in mice and humans, revealing the impact of drug exposure time on efficacy [
10]. All these systems have the common issue of difficulty retaining drugs from one-way flow and inability to assess the impact of changes in metabolite concentrations.
We focused on integrating a dialysis membrane into MPS to address these issues. The molecular selectivity of the dialysis membrane determines the size of molecules that pass through based on the Molecular Weight Cut-Off (MWCO), allowing for the retention of substances by molecular weight [
11]. Imura
et al. demonstrated that integrating dialysis membranes into MPS allows for the retention of high-molecular-weight drugs and effective waste removal [
12]. This demonstrates that drug retention by the dialysis membrane mimics continuous in vivo drug concentration changes, making it useful for evaluating drug activity. However, the developed MPS is a single tumor cell culture system, making it difficult to evaluate drug efficacy and toxicity based on metabolites through coculture with metabolically active cells. Furthermore, drug efficacy evaluation through coculture of multiple cell types requires sufficient nutrient supply for cell maintenance, and a novel system is needed to retain drugs and evaluate their efficacy.
We have developed a Dialysis Membrane-integrated Microfluidic Device (DMiMD), which maintains continuous drug concentrations using the medium change method through dialysis membranes, aiming to establish an in vitro system that mimics continuous in vivo drug concentration changes. In this study, the evaluations of the DMiMD included assessing its molecular selectivity through dialysis membranes, evaluating nutrient supply performance, and testing the efficacy of anticancer drugs using HepG2 or Upcyte liver model cells and A549 human lung cancer cells targeted by anticancer drugs. The results of the molecular selectivity evaluation confirmed that the dialysis membrane separates and retains high-molecular-weight substances while selectively supplying low-molecular-weight substances, like glucose, essential for cell culture. The results of evaluating nutrient supply performance using HepG2 and A549 cells showed that the medium change method through dialysis membranes supports cell culture similar to the conventional method. The evaluation of anticancer drug efficacy showed that the medium change through dialysis membranes allows for drug efficacy assessment under continuously changing drug concentration conditions, demonstrating that the DMiMD is an effective tool for evaluating drug efficacy and toxicity.
2. Materials and Methods
2.1. Dialysis Membrane-Integrated Microfluidic Device (DMiMD)
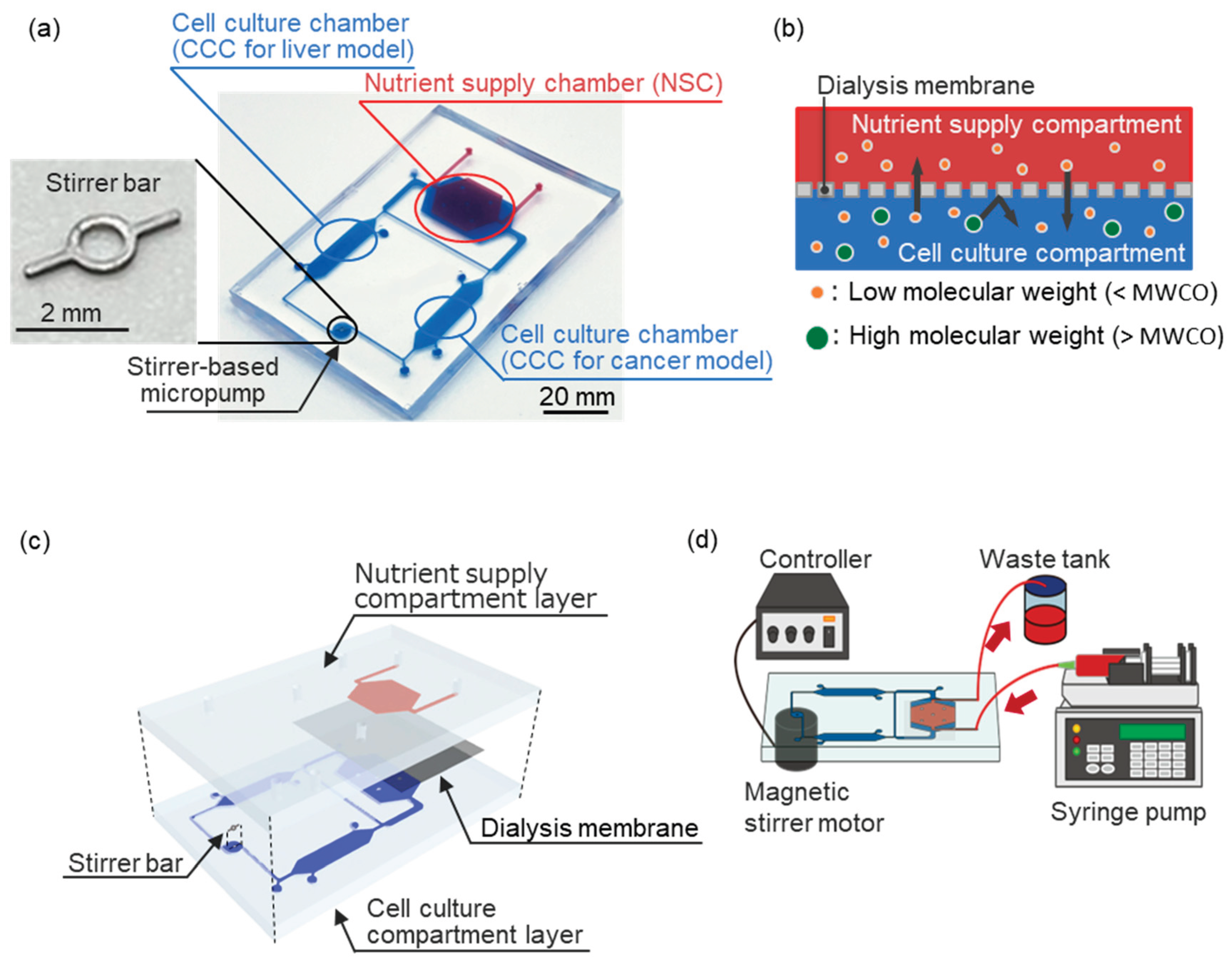
The DMiMD was based on a cell culture microfluidic device developed by Kimura
et al. [
13] and consisted of a cell culture compartment and a nutrient supply compartment (
Figure 1a). The cell culture compartment consisted of two cell culture chambers (CCC), a nutrient supply chamber (NSC), and a stirrer-based pump connected by a microfluidic channel (1.0 mm wide × 0.3 mm high). The volume of the CCC was about 50 µL. The NSC was separated into the cell culture compartment and the nutrient supply compartment by a dialysis membrane (area: 200 mm
2) (
Figure 1b). Glucose, which is an essential nutrient for cell culture, has a molecular weight of 180.16 [
14].
Therefore, the DMiMD was integrated with a dialysis membrane (F35-7935, Cellulose Tubes for Osmotic Experiments, Narika, Tokyo, Japan) having an MWCO of 3.5×10³ that efficiently permeates glucose.
Nutrients like glucose in a perfused culture medium in the NSC diffuse to the CCC by a difference in concentration via the dialysis membrane. While drugs and proteins with molecular weights larger than the MWCO are retained in the cell culture compartment by the dialysis membrane, even when the medium flows through the NSC. Therefore, the DMiMD can retain drugs in the cell culture compartment while supplying nutrients to the cultured cells. Additionally, the stirrer-based micropump developed in our previous study can generate flow in the microfluidic channel by rotating the built-in stirrer bar (3 mm long, 0.2 mm wide) in response to the rotation of a magnetic stirrer motor installed beneath the DMiMD [
15].
The DMiMD was fabricated by bonding the cell culture compartment layer chip and the nutrient supply compartment layer chip made of polydimethylpolysiloxane (PDMS, DOWSIL™SILPOT 184 W/C, Dow Toray, Tokyo, Japan) (
Figure 1c). The nutrient supply compartment layer and the cell culture compartment layer chips were molded using a mold with microchannels patterned by photolithography [
16,
17]. The microchannel height of the nutrient supply compartment and cell culture compartment layers was 0.3 mm. The nutrient supply compartment layer chip and the cell culture compartment layer chip were bonded across the dialysis membrane. The dialysis membrane was silanized to ensure strong chemical bonding with PDMS [
18]. The nutrient supply compartment layer chip and the cell culture compartment layer chip were oxygen plasma-treated at the bonding surface using a plasma cleaner and then bonded. A stainless-steel stirrer bar was used for the stirrer-based micropump in the DMiMD. A different version of the device was also fabricated with a tissue culture-treated dish plate (I3020-100, AGC TECHNO GLASS, Shizuoka, Japan) that was processed using a laser machine (zing24, Laser Connect, Tokyo, Japan) on the bottom of the CCC for culturing cells with weak adhesion. The DMiMD and the processed plate were bonded with double-sided tape (760H #25, Teraoka Seisakusho, Tokyo, Japan).
2.2. Cell Culture
HepG2 (JCRB1054, JCRB Cell Bank, Osaka, Japan) and Upcyte (KHE001-10-03, Upcyte Technologies, Hamburg, Germany) were used as liver model cells, and A549 (RCB0098, RIKEN Bio Resource Research Center, Ibaraki, Japan) were used as human lung cancer cells. These cells were cultured in an incubator at 37°C with 5% CO2. HepG2 and A549 cells were cultured in Dulbecco's Modified Eagle Medium supplemented with 10% fetal bovine serum (FBS, Bio West, Tokyo, Japan), 1% non-essential amino acids (NEAA, 11140–050, Thermo Fisher Scientific-JP, Tokyo, Japan), and 1% Antibiotic-Antimycotic solution (161–23181, FUJIFILM Wako, Osaka, Japan). Upcyte cells were cultured in Upcyte Hepatocyte High Performance Medium (MHE003, Upcyte Technologies, Hamburg, Germany) supplemented with 10% FBS, 1% NEAA, and 1% Antibiotic-Antimycotic solution.
Before cell seeding, the cell culture compartment was coated with collagen I-P (170323, Nitta Gelatin, Osaka, Japan). HepG2 or Upcyte cells were seeded at a 1.0×10⁵ cells/cm² density in the CCC for the liver model. HepG2 cells were pretreated with 50 µM Rifampicin for 3 days to induce CYP3A4 during the evaluation of anticancer drug efficacy [
19,
20]. After 3 days of confluence in liver model cells, A549 cells were seeded at a 5.0×10
4 cells/cm
2 density in the CCC for the cancer model.
The medium in the cell culture compartment was unchanged, and nutrients were supplied from the NSC via the dialysis membrane. The medium in NSC was pumped at a flow rate of 1.0 µL/min using a syringe pump (EUDC24B8, Minato Concept) (
Figure 1d). The stirrer-based micropump was driven by the magnetic stirrer motor installed beneath the DMiMD, with a 3.0 µL/min flow rate.
2.3. Cell Density Evaluation
Cell viability was evaluated by a cell staining method. Cells in the DMiMD were stained with Hoechst 33342 (346-07951, DOJINDO LABORATORIES, Kumamoto, Japan) for total nuclei and PI (341-07881, DOJINDO LABORATORIES, Kumamoto, Japan) for dead nuclei. The total and dead cell numbers were determined from fluorescence images obtained using a fluorescent microscope. The cell density of live cells was calculated from the difference between the total and dead cell numbers.
2.4. Quantification of Fluorescent Substances
The amount of fluorescent substance in the CCC of the cell culture compartment was quantified from fluorescence images obtained using a research stereomicroscope (SMZ-25, NIKON CORPORATION, Tokyo, Japan). Solutions with varying fluorescent concentrations were placed in the CCC, and calibration curves of fluorescent concentrations and brightness values were generated from fluorescence observation. The brightness values of the fluorescence images of the CCC were measured, and the concentration of fluorescent substance was calculated based on the obtained brightness values using the calibration curve.
2.5. Statistical Analysis
All values are expressed as the mean ± SD of at least three independent experiments. Statistical analysis was performed using GraphPad Prism (version 10.3.0). Data was evaluated using the Tukey-Kramer test, and statistical significance was determined at p < 0.05.
3. Results and Discussion
3.1. Evaluation of the Molecular Selectivity Using the DMiMD
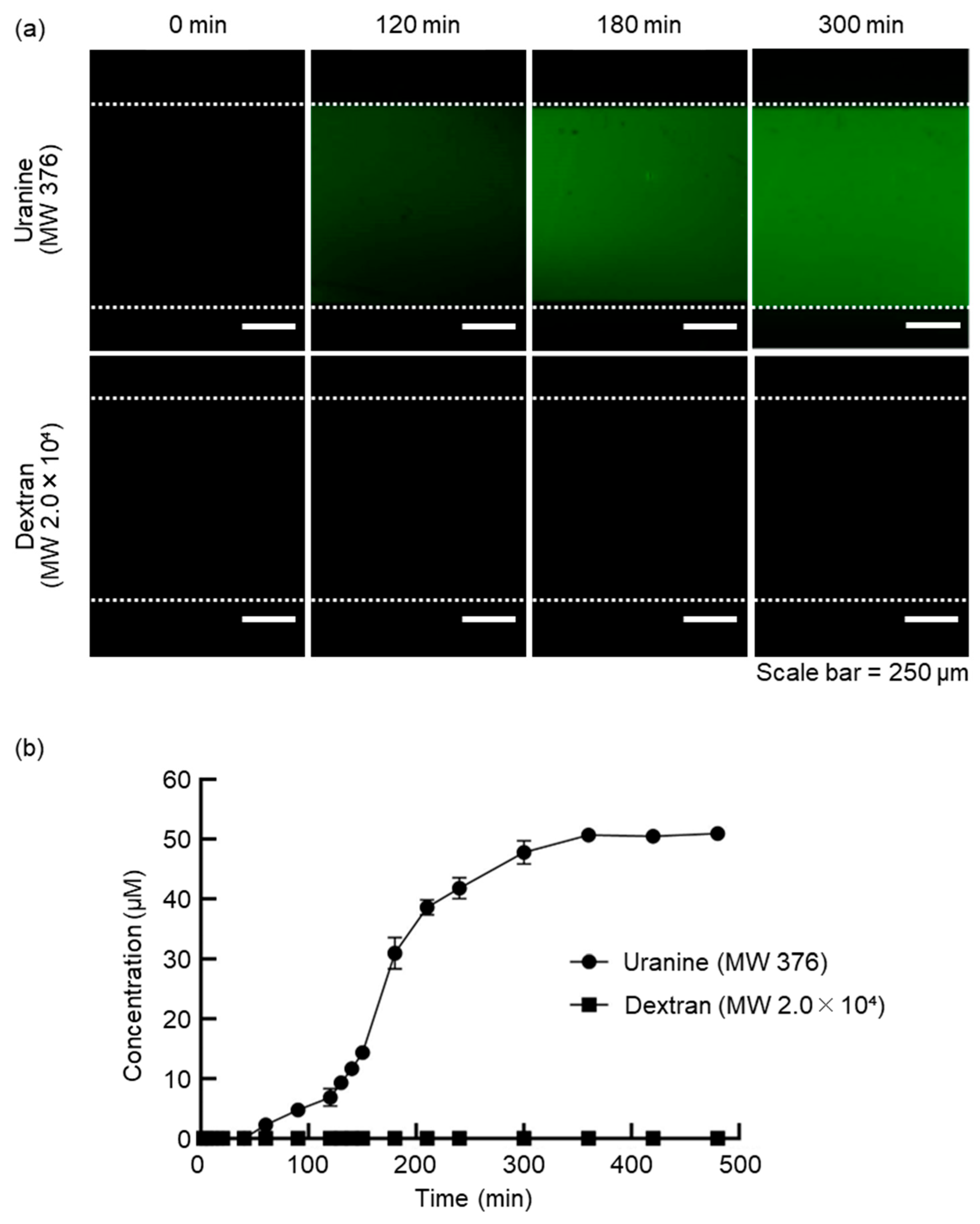
To investigate the molecular selectivity of the dialysis membrane in the DMiMD, we evaluated the changes in substance concentrations in the cell culture compartment when substances of different molecular weights were introduced into the NSC. When fresh medium was introduced into the NSC using a syringe pump in the DMiMD, substances with a molecular weight smaller than the MWCO of the dialysis membrane diffused into the cell culture compartment through the membrane. Additionally, substances with a molecular weight larger than the MWCO of the dialysis membrane are retained and do not permeate through it. The DMiMD was integrated with the dialysis membrane with an MWCO of 3.5×103, allowing sufficient glucose permeation (Molecular weight: 180.16), an essential nutrient for cell culture. In this experiment, medium containing 50 µM Uranine (Molecular weight: 376) (F0096, Tokyo Chemical Industry, Tokyo, Japan) and 50 µM Dextran (Molecular weight: 2.0×10⁴) (FD20S, Sigma-Aldrich Japan, Tokyo, Japan) was introduced to the NSC using the syringe pump.
In the CCC of the cell culture compartment, the fluorescence of Uranine increased over time. In contrast, the fluorescence of Dextran was hardly observed (
Figure 2a). As a result of calculating the changes in fluorescence substance concentrations, the concentration of Uranine in the CCC increased over time. The concentration of Dextran in the CCC remained almost zero until the end of the experiment (
Figure 2b). After 360 min, the concentration of Uranine in the CCC became 50 µM, the same as that of the fluorescent substance solution introduced to the NSC.
The molecular weight of Uranine is smaller than the MWCO of the dialysis membrane, while the molecular weight of Dextran is larger. The temporal concentration changes of Uranine and Dextran in the CCC indicate that Uranine permeated and diffused from the NSC to the cell culture compartment, while Dextran remained in the NSC without permeating the membrane. As a result, we demonstrated that the DMiMD with molecular selectivity enables the permeation and retention of molecules. Glucose is an essential nutrient for cell culture. The molecular weight of glucose is smaller than that of Uranine. Therefore, glucose permeates the dialysis membrane in the DMiMD, enabling continuous nutrient supply to the cell culture compartment through the membrane by pumping fresh medium to the NSC.
3.2. Evaluation of the Nutrient Supply Performance Using the DMiMD
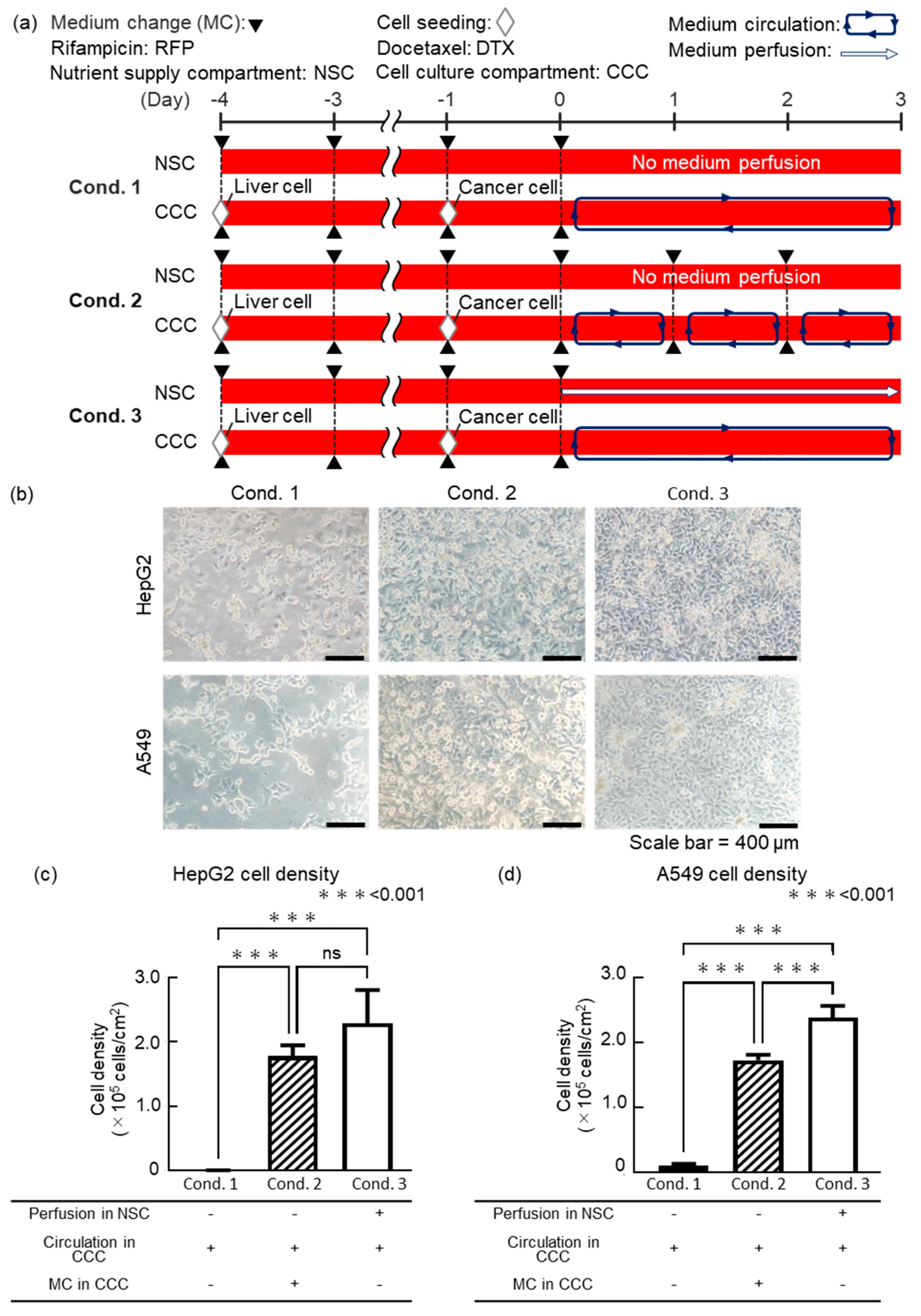
We evaluated the cell viability under different medium change methods when coculturing liver and lung cancer model cells using the DMiMD to investigate whether cell culture is possible with nutrient supply through the dialysis membrane. In this experiment, HepG2 as liver model cells and A549 as lung cancer model cells were cocultured in the DMiMD. DMEM high glucose (043-30085, FUJIFILM Wako, Osaka, Japan) was used as the medium. The following three experimental conditions were performed. Condition 1 (cond. 1) showed no medium perfusion in the NSC, without medium change in the cell culture compartment medium as the control. Condition 2 (cond. 2) was no medium perfusion in the NSC with medium change every 24 h in the cell culture compartment. Condition 3 (cond. 3) was medium perfusion in the NSC without medium change in the cell culture compartment. HepG2 cells were pre-cultured for 4 days, and A549 cells for 1 day. After that, the medium in the cell culture compartment was perfused for 3 days using the stirrer-based micropump (
Figure 3a).
In cond.1, HepG2 and A549 cells were sparse on day 3 of coculture at the endpoint. In cond. 2, HepG2 and A549 cells were confluent, but their size was slightly larger. In cond. 3, HepG2 and A549 cells were confluent and showed a pavement-like morphology (
Figure 3b). The cell density of both HepG2 and A549 was lowest in cond. 1, which was the control (
Figure 3c,d). There was no significant difference in the cell density of HepG2 between cond. 2 and cond. 3, although cond. 3 showed a higher trend. The cell density of A549 was significantly higher in cond. 3 than in cond. 2.
The glucose consumption rate of HepG2 is 37 µg/(h·10
-6 cells) [
20], while the glucose consumption rate of A549 is 27 µg/(h·10
-6 cells) [
21]. In this experiment, assuming that HepG2 and A549 cells would not proliferate from the seeding density, glucose depletion was expected after about 53 h of coculture. In cond. 1, the extremely low cell density of both HepG2 and A549 suggested glucose depletion in the medium. In cond. 2, glucose was supplied above the predicted consumption levels 270 µg/((HepG2 1.0×10
5 cells + A549 5.0×10
4 cells)·day) of HepG2 and A549 by changing the medium in the CCC every 24 hours. However, in cond. 2, the cell density of HepG2 and A549 was lower compared to cond. 3. This suggests that the glucose consumption by cells in the cell culture compartment increased with cell proliferation. As a result, the glucose level in the cell culture compartment may not have been sufficient for cell culture. In cond. 3, fresh medium was pumped to the NSC, supplying glucose to the cell culture compartment through the dialysis membrane. This condition may have contributed to the healthy culture of HepG2 and A549 cells. Based on these findings, fresh medium was pumped to the NSC in the DMiMD, supplying sufficient glucose to the cell culture compartment to maintain the cells. This demonstrated that nutrient supply equivalent to or better than the conventional method of regularly changing the entire medium is possible.
3.3. The Evaluation of Anticancer Drug Efficacy Using the DMiMD
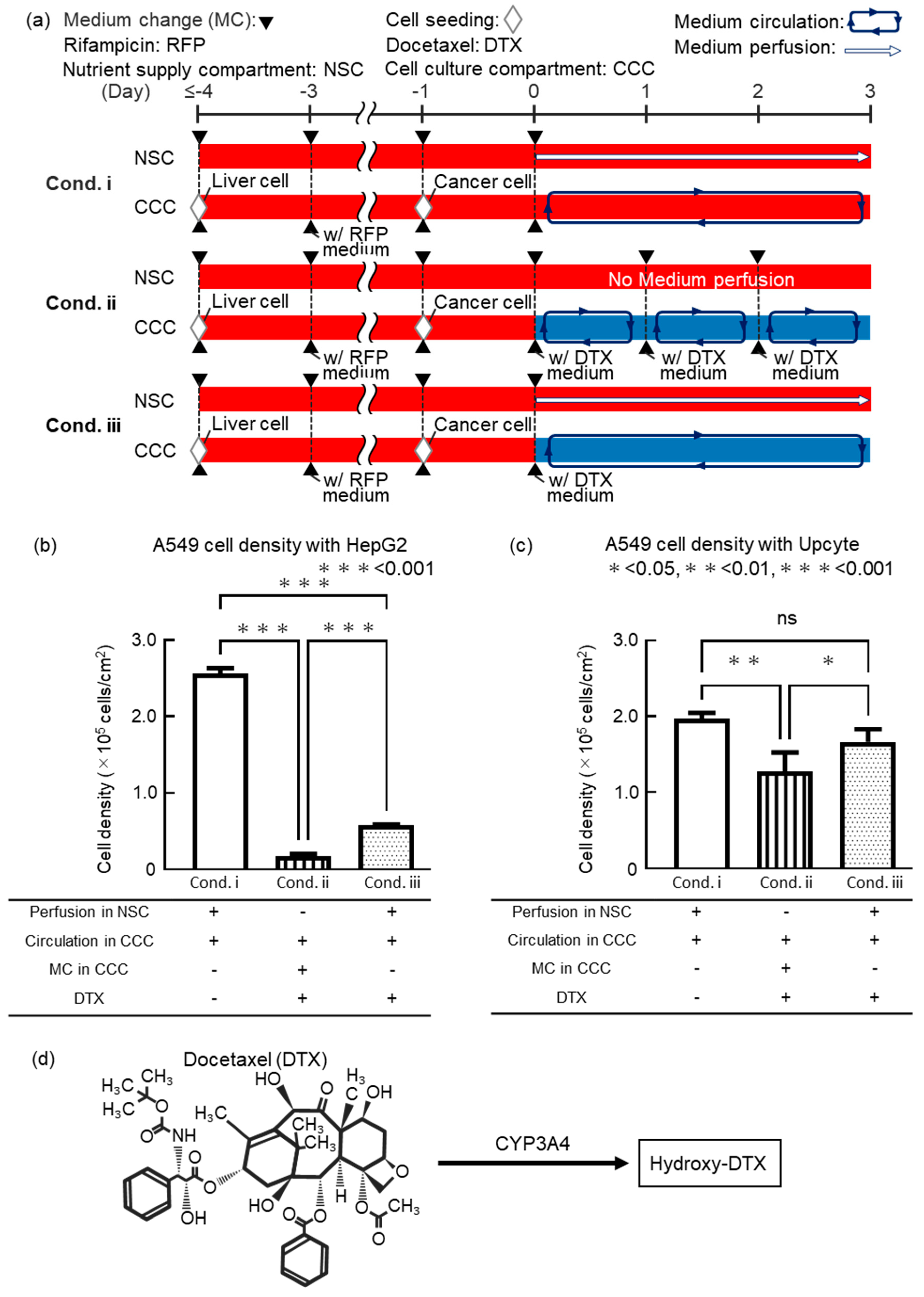
We conducted a drug efficacy test with an anticancer drug to validate the usefulness of the DMiMD. In this experiment, liver model cells and anticancer drug-targeted cells were cocultured in the DMiMD and exposed to the anticancer drug Docetaxel (DTX, 047-31281, FUJIFILM Wako, Osaka, Japan). HepG2 or Upcyte were used as liver model cells, and A549 was used as anticancer drug-targeted cells. When coculturing Upcyte and A549, the DMiMD with a tissue culture-treated dish plate was used due to the weak adhesion of Upcyte. The following three experimental conditions were performed. Condition i (cond. i) showed medium perfusion in the NSC without DTX as the negative control. Condition ii (cond. ii) was DTX medium change in the cell culture compartment. Condition iii (cond. iii) was fresh medium perfusion in the NSC with no DTX medium change in the cell culture compartment. Exposure time for 0.1 μM DTX was 3 days (
Figure 4a).
In the case of HepG2, the cell density of A549 in cond. ii_HepG2 and cond. iii_HepG2 was significantly lower than that in cond. i_HepG2 (negative control). A549 cell density in cond. iii_HepG2 was significantly higher than in cond. ii_HepG2 (
Figure 4b). In the case of Upcyte, the cell density of A549 in cond. ii_Upcyte was significantly lower than that in cond. i_Upcyte. However, the cell density of A549 in cond. iii_Upcyte showed no significant difference compared to cond. i_Upcyte (negative control). The cell density of A549 in cond. iii_Upcyte was significantly higher than that in cond. ii_Upcyte (
Figure 4c). The trend that the cell density of A549 in cond. iii was higher than in cond. ii was consistent for both HepG2 and Upcyte. However, there was a clear difference in the cell density of A549 between cond. ii and cond. iii, with lower cell density in the case of HepG2 and higher in the case of Upcyte.
DTX is a cytotoxic drug with potent anti-mitotic activity against a wide range of cancers, including lung cancer [
22], and promotes the polymerization of tubulin in cancer cells and inhibits the depolymerization of microtubules. Tubulin polymerized by DTX forms stable microtubules, but the microtubules are prevented from depolymerizing, leading to the formation of abnormal microtubule bundles inside the cell. The abnormal formation of microtubules inside the cell inhibits normal division and halts cell mitosis. As a result, apoptosis is triggered [
23,
24]. DTX is metabolized by hepatic CYP3A4 into hydroxylated derivatives, resulting in detoxification (
Figure 4d) [
25,
26]. Furthermore, DTX is known to bind to albumin, a high molecular weight protein in the blood [
27]. DTX bound to albumin is predicted not to permeate through the dialysis membrane, as its molecular weight exceeds the MWCO of the membrane. Therefore, in cond. iii, even though glucose was supplied from the NSC to the cell culture compartment, DTX was expected to remain in the cell culture compartment. When the liver model cell was HepG2, the lower cell density of A549 in cond. ii_HepG2 and cond. iii_HepG2 compared to the control cond. i_HepG2 suggests that DTX remained in the cell culture compartment and exhibited efficacy against A549.
When the liver model cell was Upcyte, the cell density of A549 in both cond. ii and cond. iii was higher than when HepG2 was used. (
Figure 4b,c). This is likely because Upcyte showed 45 to 143 times higher CYP3A4 activity than HepG2, leading to enhanced detoxification of DTX [
28]. In this experiment, although HepG2 was exposed to Rifampicin to induce CYP3A4, the original HepG2 cells have low CYP3A4 expression, so even after exposure to Rifampicin, they may not produce enough CYP3A4 to detoxify DTX. Moreover, the cell density of A549 in cond. iii_Upcyte was significantly higher than in cond. ii_Upcyte and was equivalent to that in the negative control (cond. i_Upcyte), which was not exposed to DTX. This is likely because the area under the concentration-time curve (AUC) of DTX differs between cond. ii and cond. iii. The AUC of DTX for 3days differs between cond. ii, in which the medium in the cell culture compartment was refreshed every 24 hours, and cond. iii, in which nutrients are supplied through a dialysis membrane. That is, the AUC in cond. ii is considered to be larger than that in cond. iii. As a result, it is considered that sufficient drug efficacy was not observed in cond. iii_Upcyte because Upcyte metabolized DTX. This is due to the reproduction of continuous drug concentration changes similar to those in vivo. In conventional cell culture methods, drug concentrations are reset with each medium change, making it difficult to reproduce in vivo conditions and possibly leading to inaccurate drug efficacy evaluation.
Based on these findings, the DMiMD was shown to retain drugs in the cell culture compartment by the medium change method through dialysis membranes, enabling the evaluation of anticancer drug efficacy under continuously changing drug concentration conditions. We concluded that the DMiMD is a useful tool for drug efficacy testing.
4. Conclusions
In this study, we developed the DMiMD as a novel in vitro drug efficacy test system to achieve drug concentration changes similar to in vivo conditions. The DMiMD integrated dialysis membrane between the cell culture compartments and the NSC, allowing exposure to an anticancer drug under concentration changes similar to in vivo conditions by nutrient supply via the dialysis membrane. The molecular selectivity of the DMiMD was evaluated using substances of different molecular weights. The results showed that the DMiMD had a function of selective permeation and retention of substances based on molecular weight. Comparing nutrient supply methods in the coculture of HepG2 and A549 cells showed that introducing fresh medium to the NSC can supply sufficient glucose to the cultured cells through the dialysis membrane. The results of coculturing cells for metabolism and cells for the target of drugs and exposure to the anticancer drug DTX showed that the DMiMD functions as an in vitro drug efficacy testing system. We confirmed that the nutrient supply methods through a dialysis membrane can be used to evaluate the efficacy of anticancer drugs under continuously changing drug concentrations. These results indicate that the DMiMD functions as an in vitro drug efficacy testing system, which provides continuous glucose supply to cultured cells and achieves drug concentration changes similar to in vivo conditions. The DMiMD, which realizes the nutrient supply method for retaining anticancer drugs, are expected to be an innovative tool in the fields of drug discovery and biology because of its functions as an in vitro drug efficacy testing system similar to in vivo conditions.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Funding
This work was partially supported by JSPS KAKENHI (Grant numbers JP18H01849 and JP24K01321), and the Japanese Agency for Medical Research and Development (Grant numbers JP17be0304201 and JP22be1004201).
Data Availability Statement
The original contributions presented in this study are included in the article/
Supplementary Materials. Further inquiries can be directed to the corresponding author.
Acknowledgments
We would like to thank the Tokai University Imaging Center for Advanced Research (TICAR) for their technical assistance.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- van Berlo, D.; van de Steeg, E.; Amirabadi, H.E.; Masereeuw, R. The Potential of Multi-Organ-on-Chip Models for Assessment of Drug Disposition as Alternative to Animal Testing. Current Opinion in Toxicology 2021, 27, 8–17. [Google Scholar] [CrossRef]
- Breslin, S.; O’Driscoll, L. Three-Dimensional Cell Culture: The Missing Link in Drug Discovery. Drug Discovery Today 2013, 18, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Sakai, Y.; Fujii, T. Organ/Body-on-a-Chip Based on Microfluidic Technology for Drug Discovery. Drug Metabolism and Pharmacokinetics 2018, 33, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Nishikawa, M.; Kutsuzawa, N.; Tokito, F.; Kobayashi, T.; Kurniawan, D.A.; Shioda, H.; Cao, W.; Shinha, K.; Nakamura, H.; et al. Advancements in Microphysiological Systems: Exploring Organoids and Organ-on-a-Chip Technologies in Drug Development -Focus on Pharmacokinetics Related Organs-. Drug Metabolism and Pharmacokinetics 2025, 60, 101046. [Google Scholar] [CrossRef]
- Ito, Y.; Kawauchi, I.; Yanagita, Y.; Sakai, Y.; Nishikawa, M.; Arakawa, H.; Kadoguchi, M.; Tamai, I.; Esashika, K.; Takahashi, J.; et al. Microphysiological Systems for Realizing Microenvironment That Mimics Human Physiology—Functional Material and Its Standardization Applied to Microfluidics. emergent mater. 2024. [CrossRef]
- Kopec, A.K.; Yokokawa, R.; Khan, N.; Horii, I.; Finley, J.E.; Bono, C.P.; Donovan, C.; Roy, J.; Harney, J.; Burdick, A.D.; et al. Microphysiological Systems in Early Stage Drug Development: Perspectives on Current Applications and Future Impact. J. Toxicol. Sci. 2021, 46, 99–114. [Google Scholar] [CrossRef]
- Kim, J.J.; Bae, M.; Cho, D. Multi-Organ Microphysiological Systems Targeting Specific Organs for Recapitulating Disease Phenotypes via Organ Crosstalk. Small Science 2024, 4, 2400314. [Google Scholar] [CrossRef]
- Petreus, T.; Cadogan, E.; Hughes, G.; Smith, A.; Pilla Reddy, V.; Lau, A.; O’Connor, M.J.; Critchlow, S.; Ashford, M.; Oplustil O’Connor, L. Tumour-on-Chip Microfluidic Platform for Assessment of Drug Pharmacokinetics and Treatment Response. Commun Biol 2021, 4, 1001. [Google Scholar] [CrossRef]
- Singh, D.; Deosarkar, S.P.; Cadogan, E.; Flemington, V.; Bray, A.; Zhang, J.; Reiserer, R.S.; Schaffer, D.K.; Gerken, G.B.; Britt, C.M.; et al. A Microfluidic System That Replicates Pharmacokinetic (PK) Profiles in Vitro Improves Prediction of in Vivo Efficacy in Preclinical Models. PLoS Biol 2022, 20, e3001624. [Google Scholar] [CrossRef]
- Guerrero, Y.A.; Desai, D.; Sullivan, C.; Kindt, E.; Spilker, M.E.; Maurer, T.S.; Solomon, D.E.; Bartlett, D.W. A Microfluidic Perfusion Platform for In Vitro Analysis of Drug Pharmacokinetic-Pharmacodynamic (PK-PD) Relationships. AAPS J 2020, 22, 53. [Google Scholar] [CrossRef]
- Eswari, J.S.; Naik, S. A Critical Analysis on Various Technologies and Functionalized Materials for Manufacturing Dialysis Membranes. Materials Science for Energy Technologies 2020, 3, 116–126. [Google Scholar] [CrossRef]
- Imura, Y.; Yoshimura, E.; Sato, K. Microcirculation System with a Dialysis Part for Bioassays Evaluating Anticancer Activity and Retention. Anal. Chem. 2013, 85, 1683–1688. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Ikeda, T.; Nakayama, H.; Sakai, Y.; Fujii, T. An On-Chip Small Intestine-Liver Model for Pharmacokinetic Studies. J Lab Autom 2015, 20, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Koirala, R.P.; Dawanse, S.; Pantha, N. Diffusion of Glucose in Water: A Molecular Dynamics Study. Journal of Molecular Liquids 2022, 345, 117826. [Google Scholar] [CrossRef]
- Kimura, H.; Yamamoto, T.; Sakai, H.; Sakai, Y.; Fujii, T. An Integrated Microfluidic System for Long-Term Perfusion Culture and on-Line Monitoring of Intestinal Tissue Models. Lab Chip 2008, 8, 741–746. [Google Scholar] [CrossRef]
- McDonald, J.C.; Whitesides, G.M. Poly(Dimethylsiloxane) as a Material for Fabricating Microfluidic Devices. Acc. Chem. Res. 2002, 35, 491–499. [Google Scholar] [CrossRef]
- Kim, A.A.; Kustanovich, K.; Baratian, D.; Ainla, A.; Shaali, M.; Jeffries, G.D.M.; Jesorka, A. SU-8 Free-Standing Microfluidic Probes. Biomicrofluidics 2017, 11, 014112. [Google Scholar] [CrossRef]
- Borók, A.; Laboda, K.; Bonyár, A. PDMS Bonding Technologies for Microfluidic Applications: A Review. Biosensors 2021, 11, 292. [Google Scholar] [CrossRef]
- Mazzari, A.L.D.A.; Milton, F.; Frangos, S.; Carvalho, A.C.B.; Silveira, D.; De Assis Rocha Neves, F.; Prieto, J.M. In Vitro Effects of Four Native Brazilian Medicinal Plants in CYP3A4 mRNA Gene Expression, Glutathione Levels, and P-Glycoprotein Activity. Front. Pharmacol. 2016, 7. [Google Scholar] [CrossRef]
- Usui, T.; Saitoh, Y.; Komada, F. Induction of CYP3As in HepG2 Cells by Several Drugs. Association between Induction of CYP3A4 and Expression of Glucocorticoid Receptor. Biological & Pharmaceutical Bulletin 2003, 26, 510–517. [Google Scholar] [CrossRef]
- Shankland, E.; Livesey, J.; Wiseman, R.; Krohn, K. Multinuclear NMR Studies of an Actively Dividing Artificial Tumor. Physiol Res 2002, 49–58. [Google Scholar] [CrossRef]
- Qu, M.-H.; Zeng, R.-F.; Fang, S.; Dai, Q.-S.; Li, H.-P.; Long, J.-T. Liposome-Based Co-Delivery of siRNA and Docetaxel for the Synergistic Treatment of Lung Cancer. International Journal of Pharmaceutics 2014, 474, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, A.; Martínez-Alcázar, M.P.; Durán, I.; Calvo, E.; Valenzuela, B.; Barbas, C.; García, A. Simultaneous Online SPE–HPLC–MS/MS Analysis of Docetaxel, Temsirolimus and Sirolimus in Whole Blood and Human Plasma. Journal of Chromatography B 2013, 921–922, 35–42. [CrossRef]
- Morse, D.L.; Gray, H.; Payne, C.M.; Gillies, R.J. Docetaxel Induces Cell Death through Mitotic Catastrophe in Human Breast Cancer Cells. Molecular Cancer Therapeutics 2005, 4, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Engels, F.K.; ten Tije, A.J.; Baker, S.D.; Lee, C.K.K.; Loos, W.J.; Vulto, A.G.; Verweij, J.; Sparreboom, A. Effect of Cytochrome P450 3A4 Inhibition on the Pharmacokinetics of Docetaxel. Clinical Pharmacology & Therapeutics 2004, 75, 448–454. [Google Scholar] [CrossRef]
- Rodriguez-Antona, C.; Ingelman-Sundberg, M. Cytochrome P450 Pharmacogenetics and Cancer. Oncogene 2006, 25, 1679–1691. [Google Scholar] [CrossRef]
- Loos, W. Clinical Pharmacokinetics of Unbound Docetaxel: Role of Polysorbate 80 and Serum Proteins. Clinical Pharmacology & Therapeutics 2003, 74, 364–371. [Google Scholar] [CrossRef]
- Tolosa, L.; Gómez-Lechón, M.J.; López, S.; Guzmán, C.; Castell, J.V.; Donato, M.T.; Jover, R. Human Upcyte Hepatocytes: Characterization of the Hepatic Phenotype and Evaluation for Acute and Long-Term Hepatotoxicity Routine Testing. Toxicol. Sci. 2016, 152, 214–229. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).