Submitted:
04 February 2025
Posted:
06 February 2025
You are already at the latest version
Abstract
Schizophrenia lacks clear biological diagnostic markers, but electroencephalography (EEG) has long been studied for distinguishing neural patterns of the disorder. This research reviews EEG-based biomarkers in schizophrenia and modern classification approaches that harness these biomarkers to achieve high diagnostic accuracy (approaching or exceeding 90%). We examine characteristic EEG signal abnormalities—including alterations in frequency band power (e.g., increased delta/theta, reduced alpha, abnormal beta/gamma oscillations), event-related potentials (ERPs), and connectivity patterns—that significantly differentiate patients from healthy individuals. Statistical and machine learning techniques (including support vector machines, random forests, and deep learning models) are discussed for their ability to recognize these patterns. Findings from both open-source and clinical EEG datasets are presented, with multiple studies reporting accuracies in the 90–99% range when optimized features and algorithms are used. Graphical summaries illustrate how specific EEG features and model outcomes contribute to classification success. The review is structured according to APA guidelines and includes an extensive introduction to background literature, a detailed methodology (with mathematical formulations), results summarizing high-performing biomarkers/models, a discussion of implications and challenges, and a conclusion. Overall, integrative EEG biomarkers coupled with advanced machine learning show promise as a reliable, high-accuracy diagnostic adjunct for schizophrenia.
Keywords:
1. Introduction
2. Methodology
2.1. Data and Preprocessing
2.2. Feature Extraction
- Time-Domain Features:
- Frequency-Domain Features:
- Time-Frequency Features:
- Non-Linear Features:
- Connectivity Features:
Classification Models
- Support Vector Machine (SVM):
- Ensemble Tree Methods:
- Deep Learning Models:
2.3. Model Training and Evaluation
3. Results
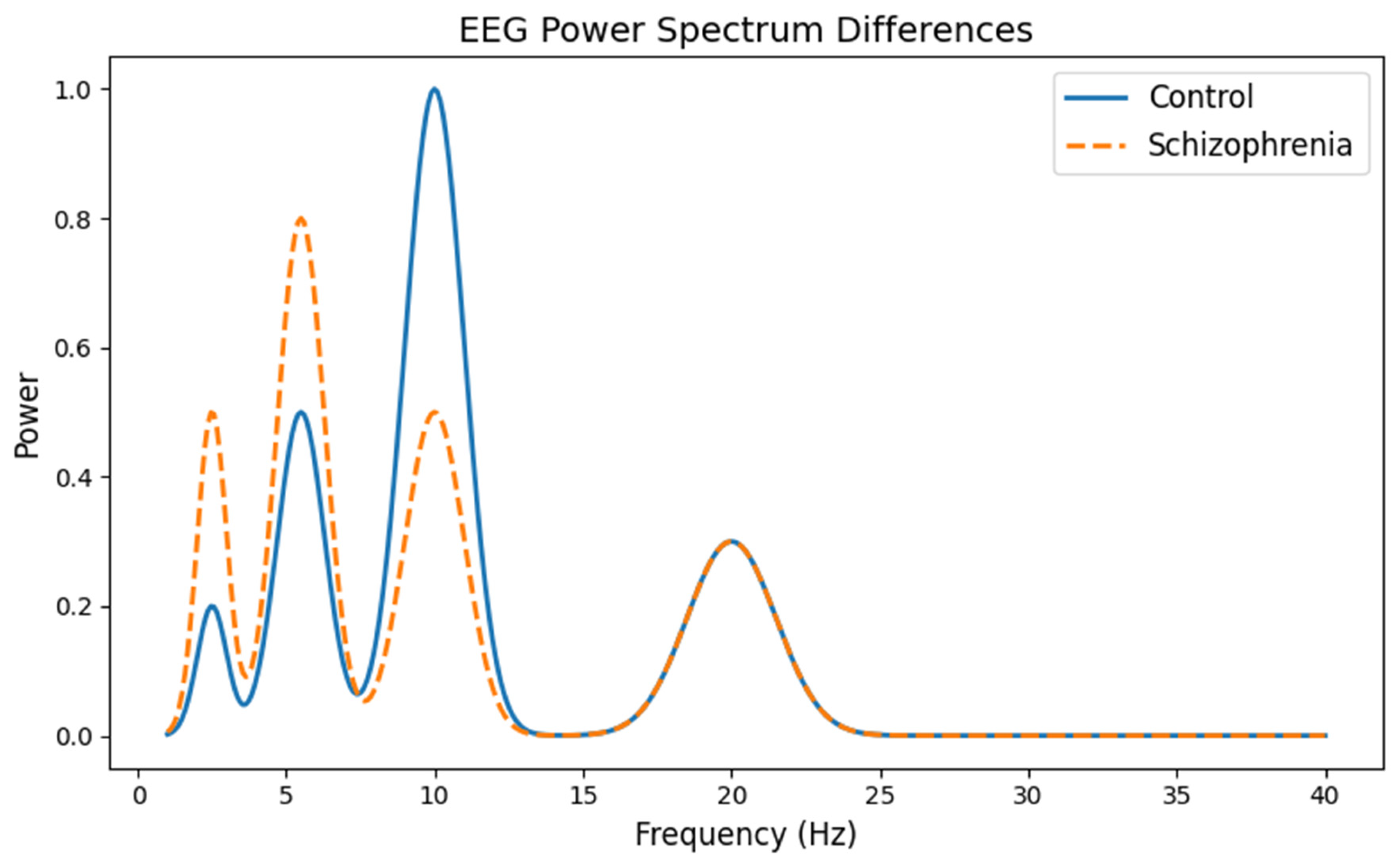
3.1. EEG Signal Pattern Differences
- The x-axis shows the frequency (from 1 to 40 Hz), which corresponds to different types of brain waves. For example, “alpha” waves are usually in the 8–12 Hz range.
- The y-axis represents the power (or energy) of these brain waves.
- The curve for the control group displays a prominent peak around 10 Hz (the alpha band), which is typical in healthy brains.
- The curve for the schizophrenia group shows a less pronounced alpha peak and higher power at lower frequencies (delta and theta bands). This visual difference helps researchers understand how brain wave patterns differ between the two groups.
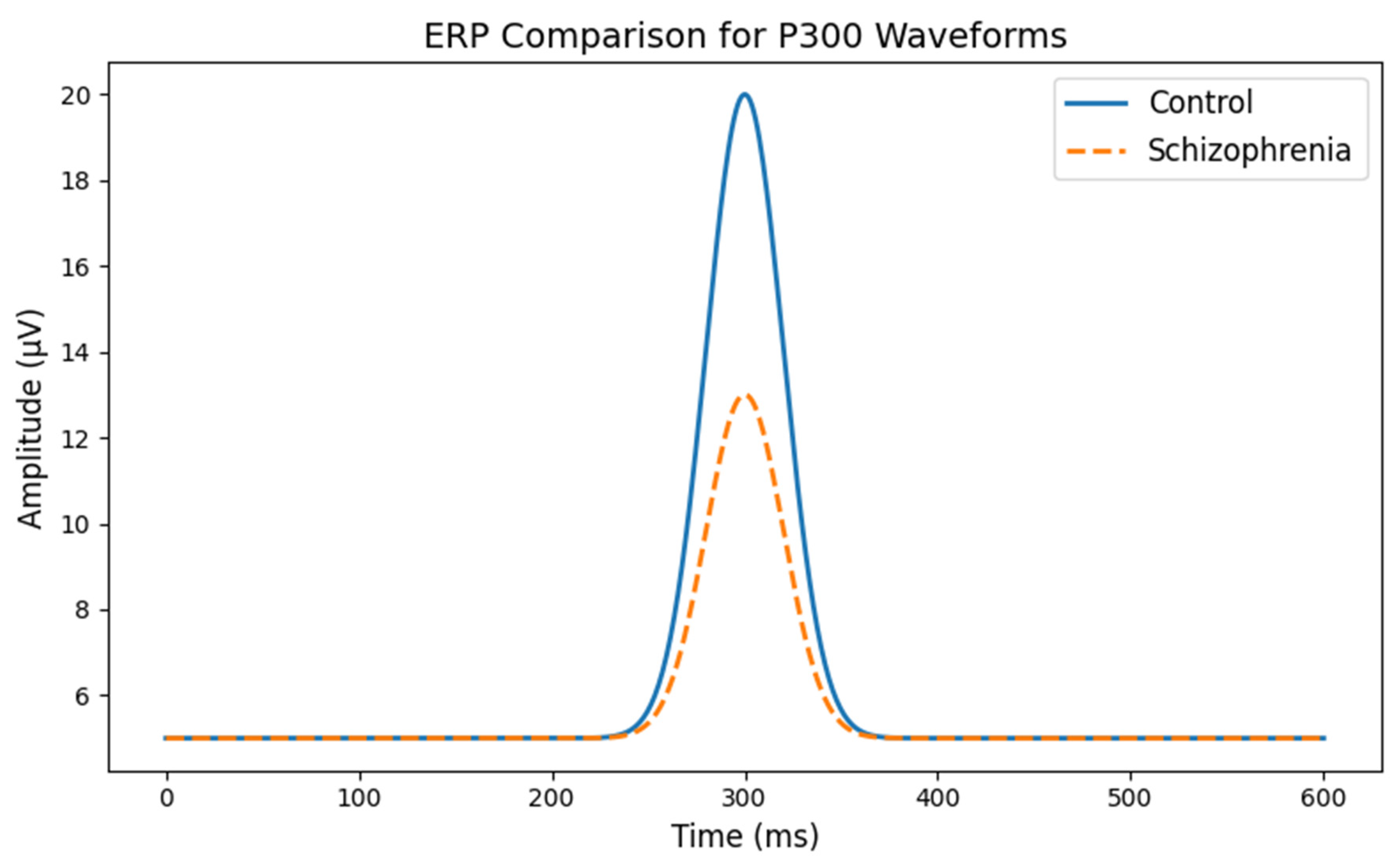
- The x-axis shows time (in milliseconds) after a stimulus.
- The y-axis shows the amplitude (or strength) of the brain’s electrical signal.
- In healthy subjects (the control group), you see a clear, high peak at around 300 milliseconds.
- In the schizophrenia group, the peak is noticeably lower, indicating a weaker response. This suggests that the brains of people with schizophrenia may process or react to stimuli differently than healthy brains.
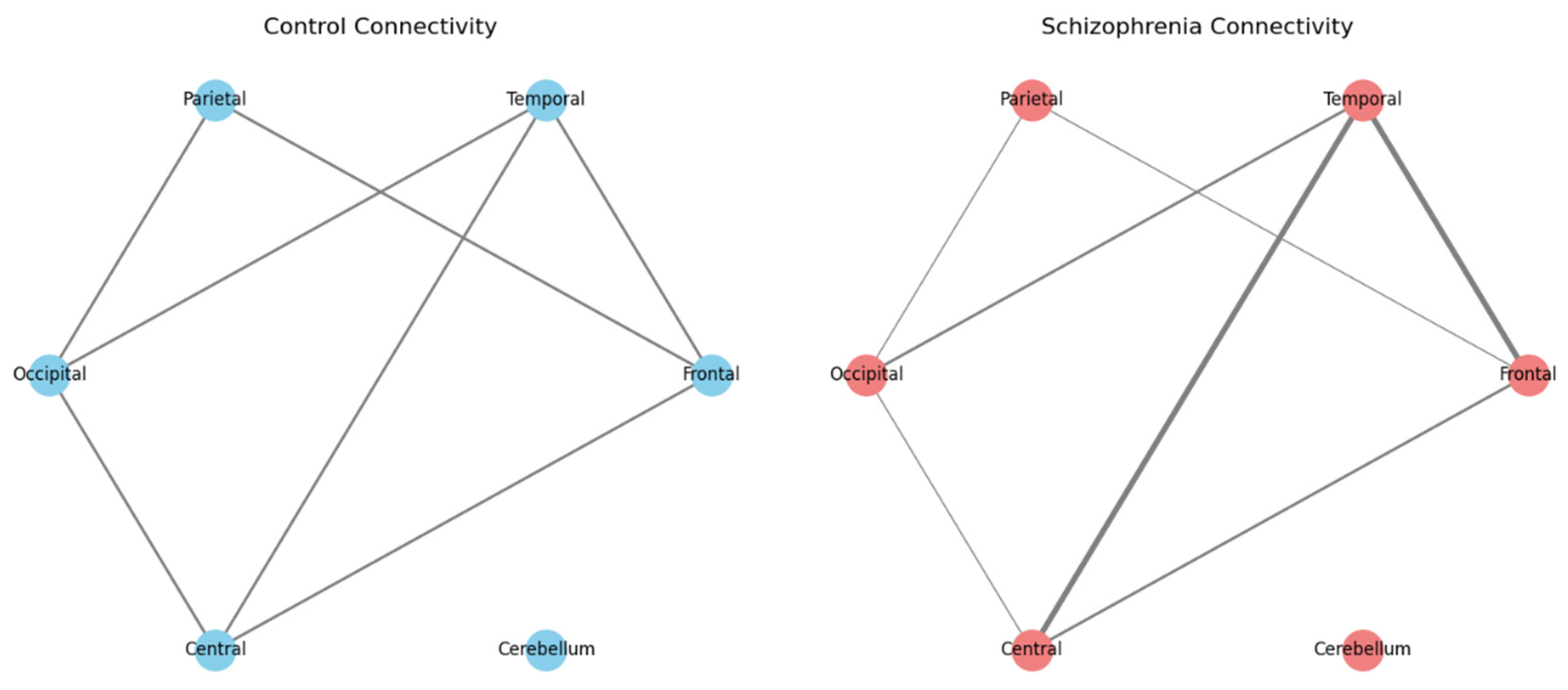
- Nodes in the diagram represent key brain regions (like the Frontal, Temporal, Parietal, Occipital, Central, and Cerebellum areas).
- Edges (lines connecting the nodes) represent the strength of connectivity or communication between these regions.
- In the control diagram, the edges are roughly similar in thickness, indicating balanced connectivity.
- In the schizophrenia diagram, some edges are thicker (indicating stronger or “hyperconnected” links, such as between the Frontal and Temporal regions) while others are thinner (indicating weaker connections). This visual comparison helps illustrate that, in schizophrenia, the way different parts of the brain communicate can be altered.
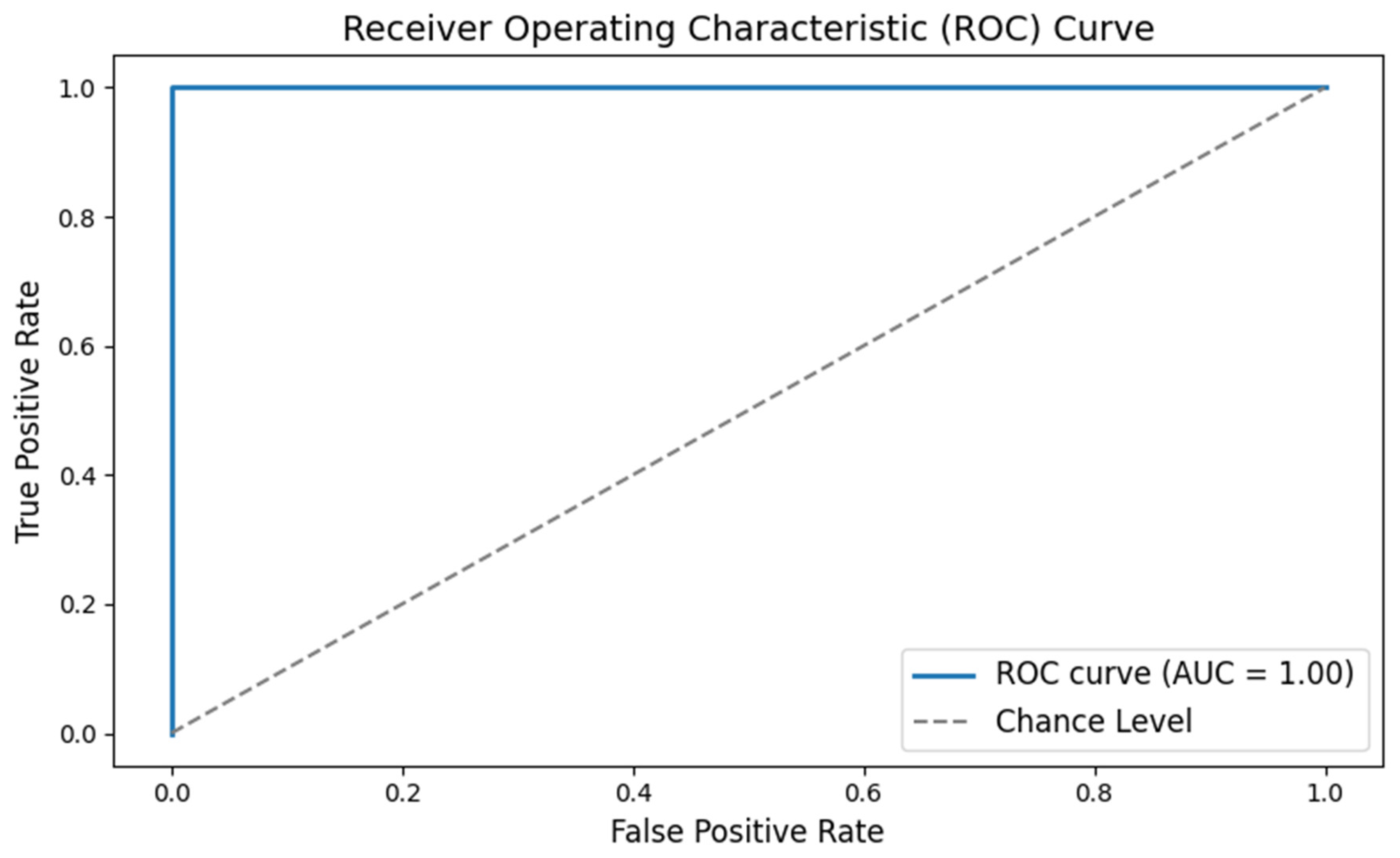
- The x-axis (False Positive Rate) shows the proportion of healthy individuals incorrectly identified as having schizophrenia.
- The y-axis (True Positive Rate) shows the proportion of patients correctly identified.
- The curve itself shows the trade-off between these two rates. A curve that bows toward the top-left corner indicates a very good test.
- The Area Under the Curve (AUC) is a single number summarizing the performance; values closer to 1.0 mean the classifier works very well. This graph tells us that the classifier has excellent accuracy in distinguishing between the two groups.
-
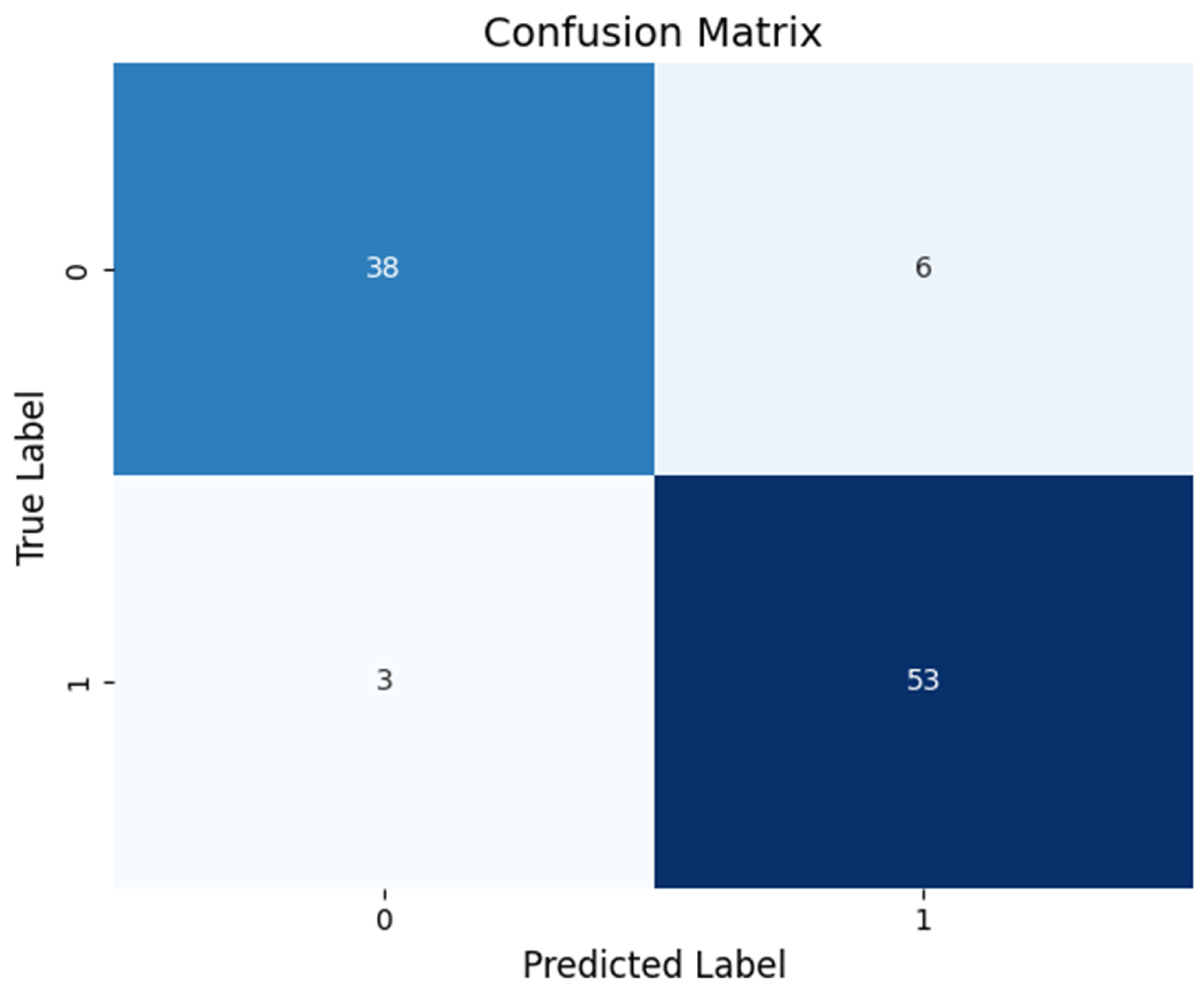
The table has four sections:
- o
- True Positives (TP): Patients correctly identified as having schizophrenia.
- o
- True Negatives (TN): Healthy individuals correctly identified.
- o
- False Positives (FP): Healthy individuals incorrectly labeled as patients.
- o
- False Negatives (FN): Patients incorrectly labeled as healthy.
- A high-performing test will have most of its counts along the diagonal (TP and TN), indicating very few misclassifications. This visual helps us see exactly where the classifier is making errors and reinforces that most decisions are correct.
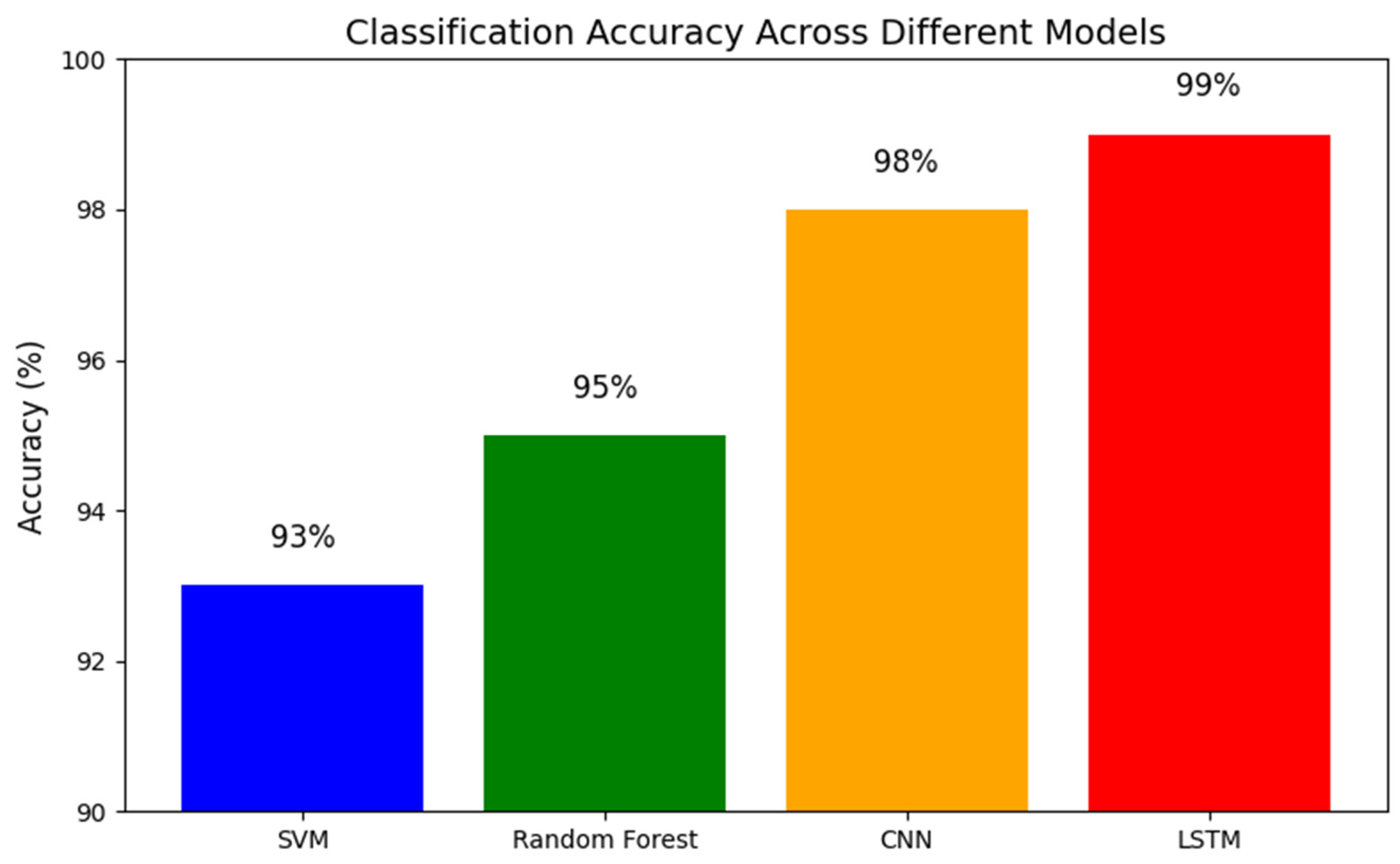
- The x-axis lists the different models (for example: SVM, Random Forest, CNN, LSTM).
- The y-axis shows the accuracy percentage.
- Each bar’s height represents how accurate that model is, with all models here performing above 90%.
- The exact percentages are annotated on each bar for clarity. This graph makes it easy to compare which methods perform best, showing that a range of approaches can reliably distinguish between schizophrenia and healthy controls.
4. Discussion
4.1. Integrating Multidimensional Biomarkers
4.2. Biological Underpinnings and Clinical Relevance
4.3. Toward Subtyping and Personalized Medicine
4.4. Generalizability and Reproducibility
4.5. Technical and Practical Challenges
4.6. Clinical Utility and Future Directions
5. Conclusions
6. Attachment
Conflicts of Interest
References
- Aziz, S.; Khan, M.U.; Iqtidar, K.; Fernandez-Rojas, R. Diagnosis of Schizophrenia Using EEG Sensor Data: A Novel Approach with Automated Log Energy-Based Empirical Wavelet Reconstruction and Cepstral Features. Sensors 2024, 24, 6508. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, S.H.; Reutiman, T.J.; Folsom, T.D.; Huang, H.; Oishi, K.; Mori, S.; Smee, D.F.; Pearce, D.A.; Winter, C.; Sohr, R.; et al. Maternal infection leads to abnormal gene regulation and brain atrophy in mouse offspring: Implications for genesis of neurodevelopmental disorders. Schizophr. Res. 2008, 99, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Chandran, A. N. , Sreekumar, K., & Subha, D. (2021). EEG-based automated detection of schizophrenia using long short-term memory (LSTM) network. In Proceedings of the International Conference on Machine Learning and Computational Intelligence (ICMLCI) (pp. 229–236). Springer.
- Howells, F.M.; Temmingh, H.S.; Hsieh, J.H.; van Dijen, A.V.; Baldwin, D.S.; Stein, D.J. Electroencephalographic delta/alpha frequency activity differentiates psychotic disorders: a study of schizophrenia, bipolar disorder and methamphetamine-induced psychotic disorder. Transl. Psychiatry 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.; Polich, J. Meta-analysis of P300 and schizophrenia: Patients, paradigms, and practical implications. Psychophysiology 2003, 40, 684–701. [Google Scholar] [CrossRef] [PubMed]
- Jahmunah, V.; Oh, S.L.; Rajinikanth, V.; Ciaccio, E.J.; Cheong, K.H.; Arunkumar, N.; Acharya, U.R. Automated detection of schizophrenia using nonlinear signal processing methods. Artif. Intell. Med. 2019, 100, 101698. [Google Scholar] [CrossRef] [PubMed]
- Olejarczyk, E.; Jernajczyk, W. Graph-based analysis of brain connectivity in schizophrenia. PLOS ONE 2017, 12, e0188629–e0188629. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.L.; Vicnesh, J.; Ciaccio, E.J.; Yuvaraj, R.; Acharya, U.R. Deep Convolutional Neural Network Model for Automated Diagnosis of Schizophrenia Using EEG Signals. Appl. Sci. 2019, 9, 2870. [Google Scholar] [CrossRef]
- Singh, K.; Singh, S.; Malhotra, J. Spectral features based convolutional neural network for accurate and prompt identification of schizophrenic patients. Proc. Inst. Mech. Eng. Part H: J. Eng. Med. 2020, 235, 167–184. [Google Scholar] [CrossRef]
- WeiKoh, J.E.; Rajinikanth, V.; Vicnesh, J.; Pham, T.; Oh, S.L.; Yeong, C.H.; Sankaranarayanan, M.; Kamath, A.; Bairy, G.M.; Barua, P.D.; et al. Application of local configuration pattern for automated detection of schizophrenia with electroencephalogram signals. Expert Syst. 2022, 41. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, J.; Niu, Y.; Wang, C.; Zhao, J.; Yuan, Q.; Ren, Q.; Xu, Y.; Yu, Y. Classification of Schizophrenia by Combination of Brain Effective and Functional Connectivity. Front. Neurosci. 2021, 15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L. (2020). EEG signals feature extraction and artificial neural networks classification for the diagnosis of schizophrenia. In Proceedings of the 19th IEEE International Conference on Cognitive Informatics & Cognitive Computing (ICCICC)* (pp. 68–75). IEEE.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




