Submitted:
24 December 2024
Posted:
25 December 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
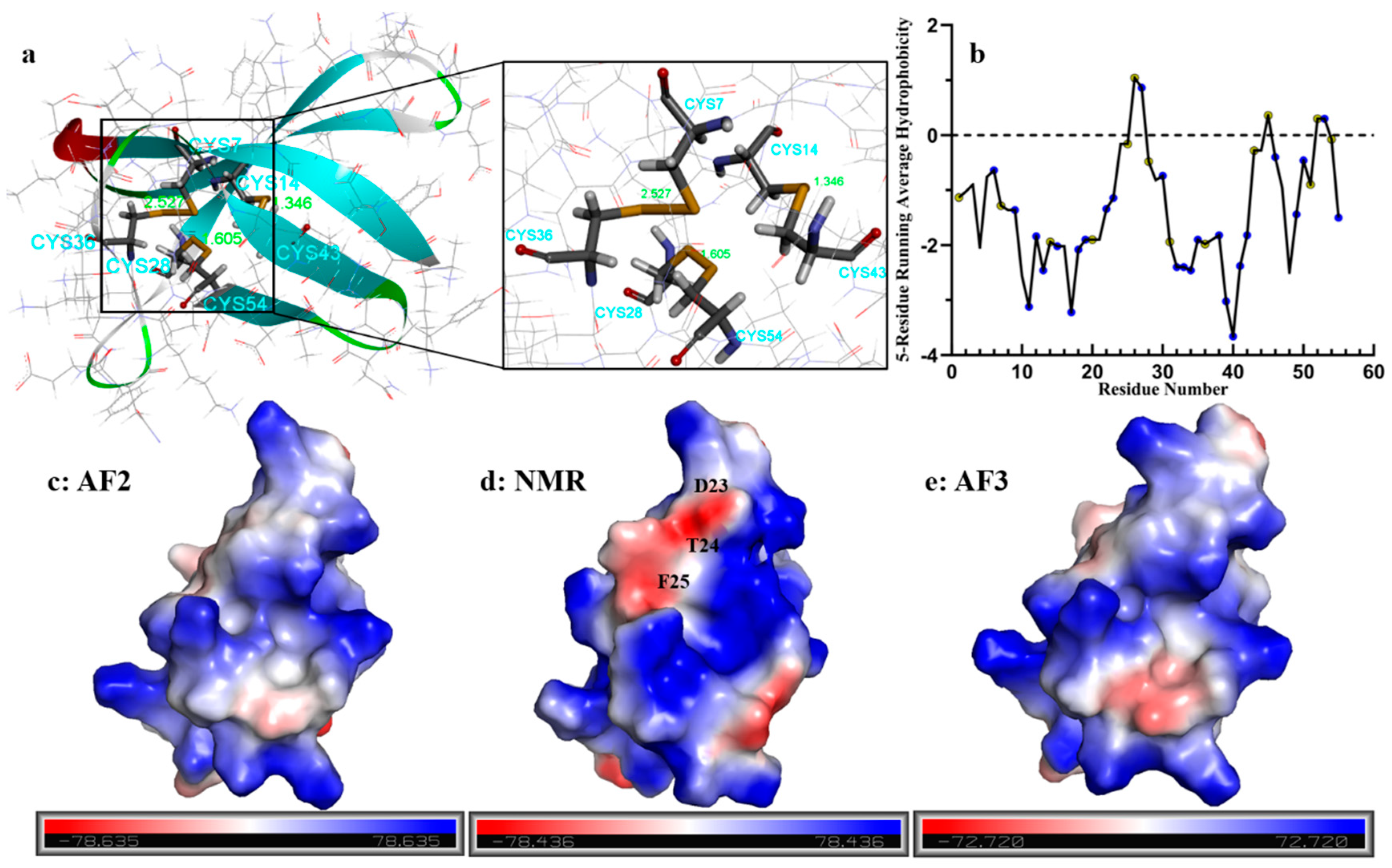
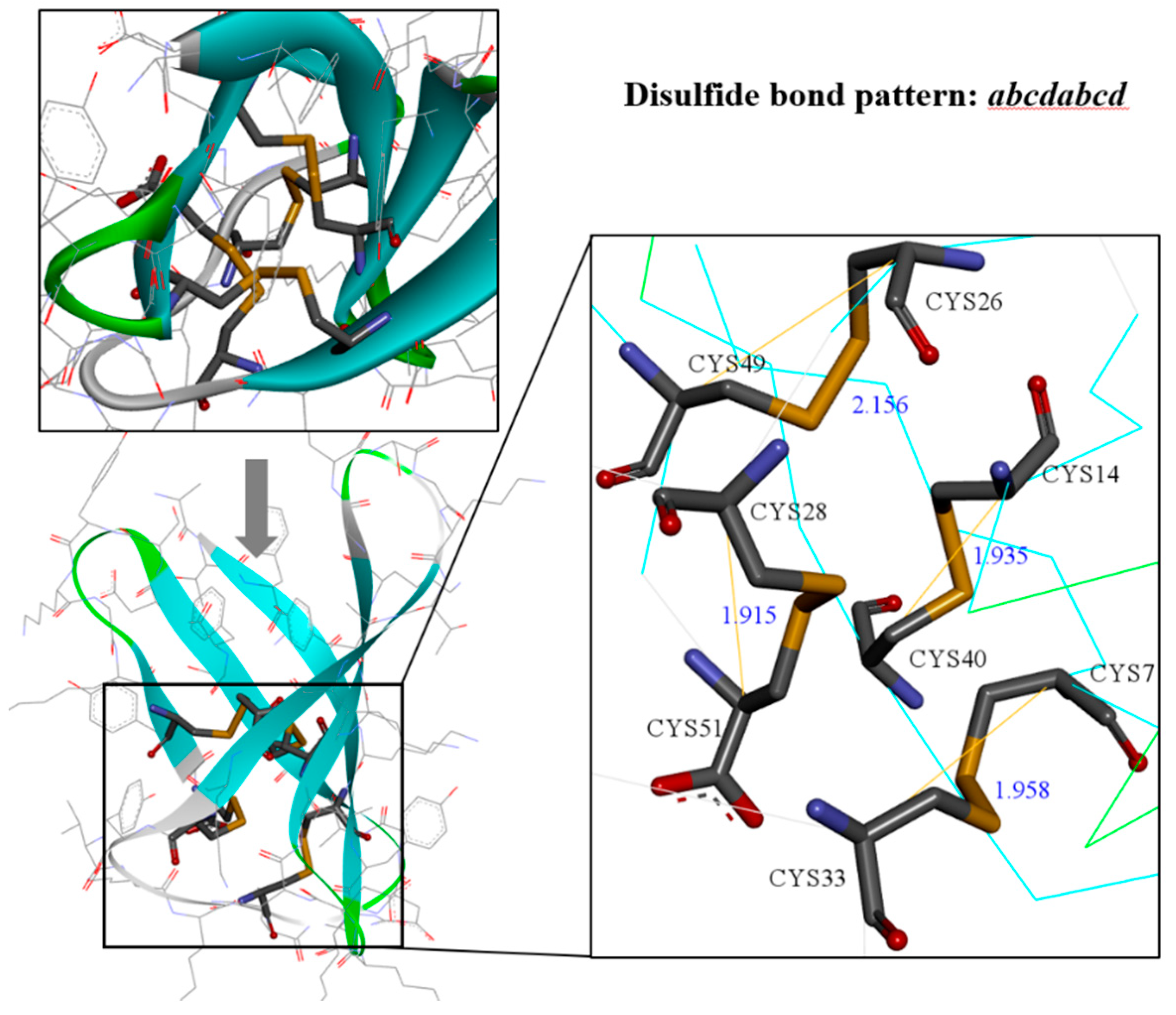
2.1. Bioinformatics Analysis of the Primary Structure of AFPs
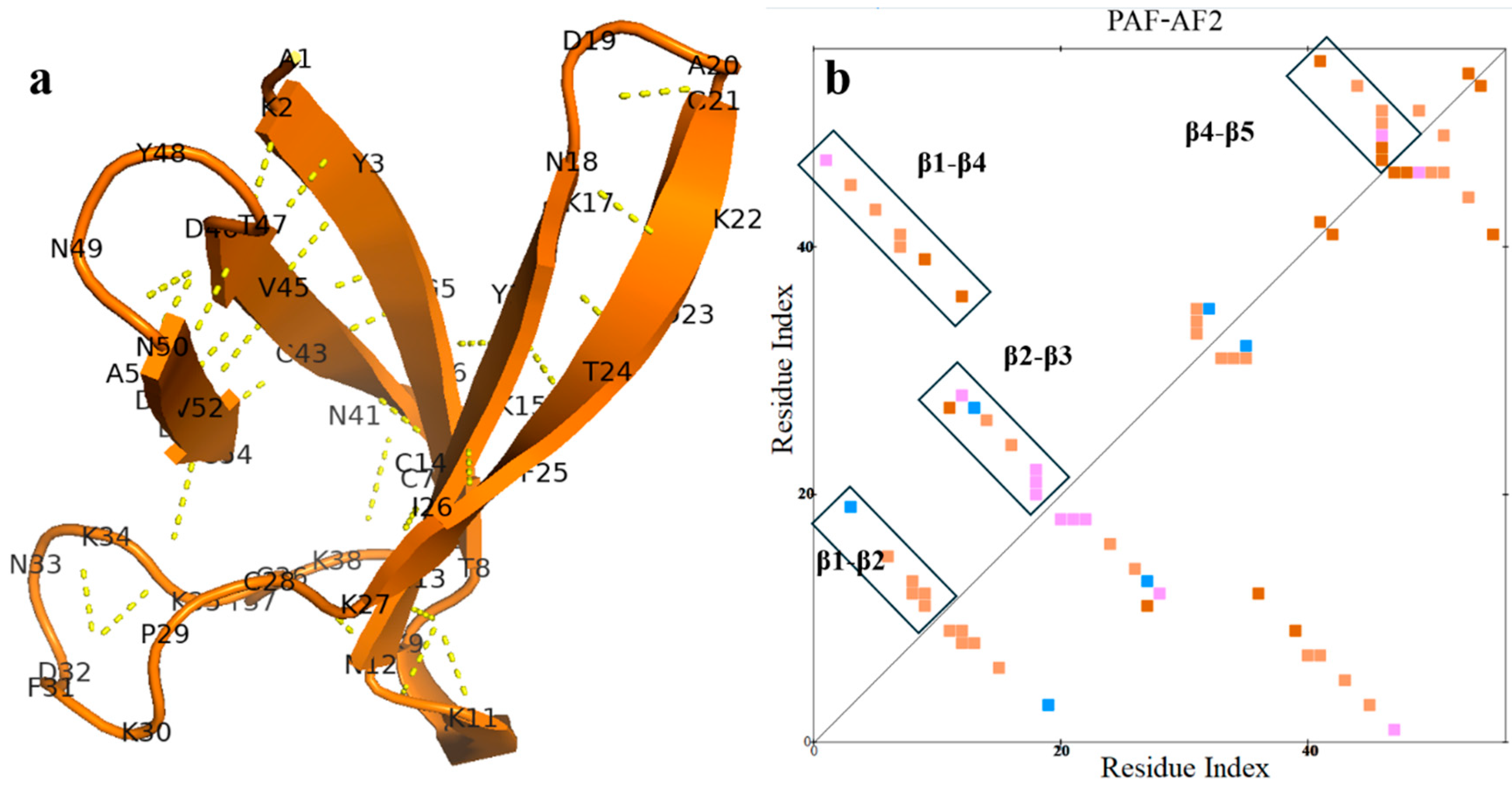
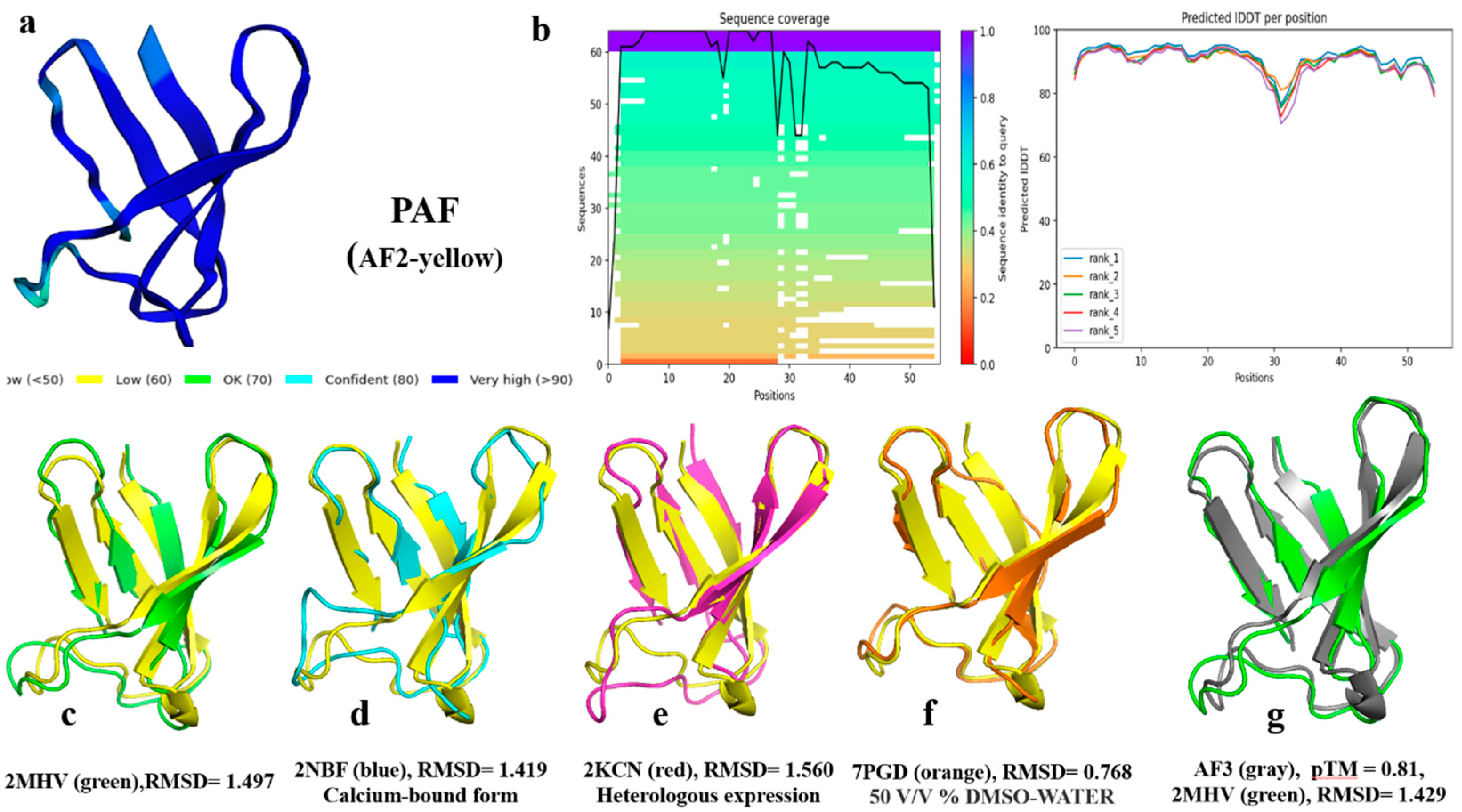
2.2. AlphaFold2 Prediction of PAF Structural Model
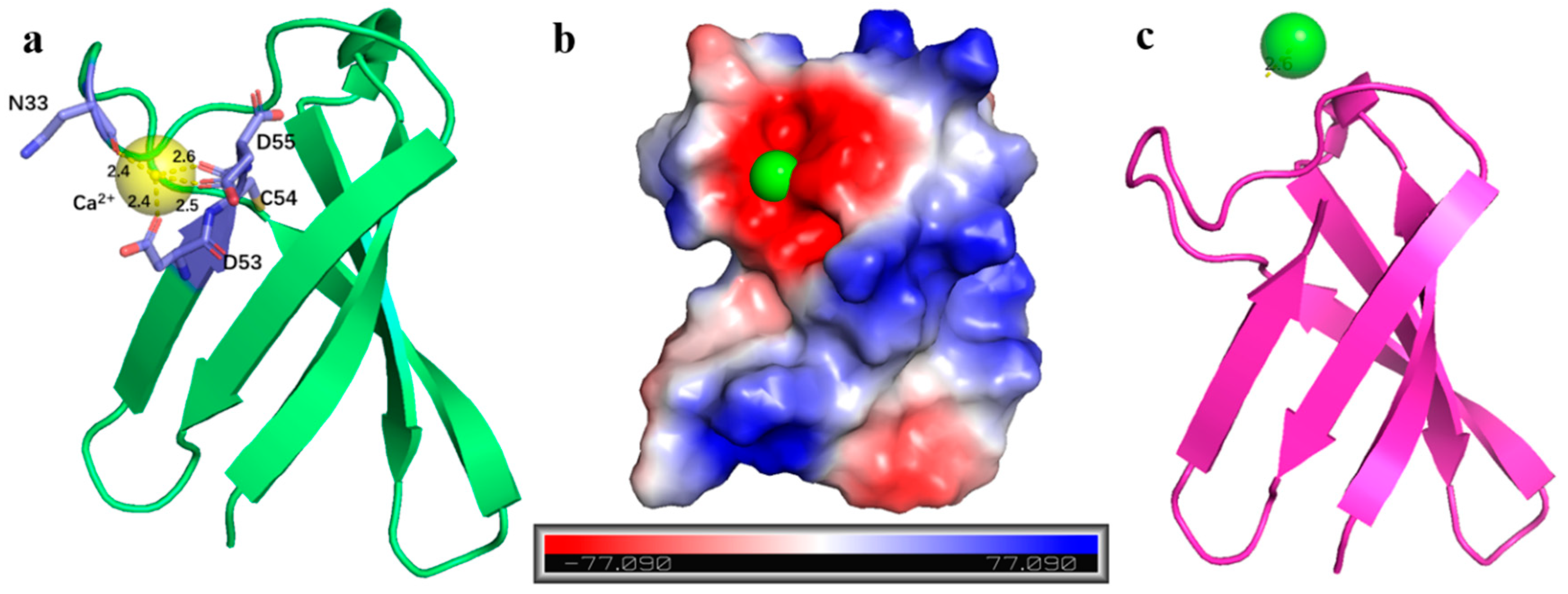
2.3. AlphaFold3 Prediction of PAF-Metal Ion Complex Structure Models
2.4. MolProbity Structural Evaluation Analysis
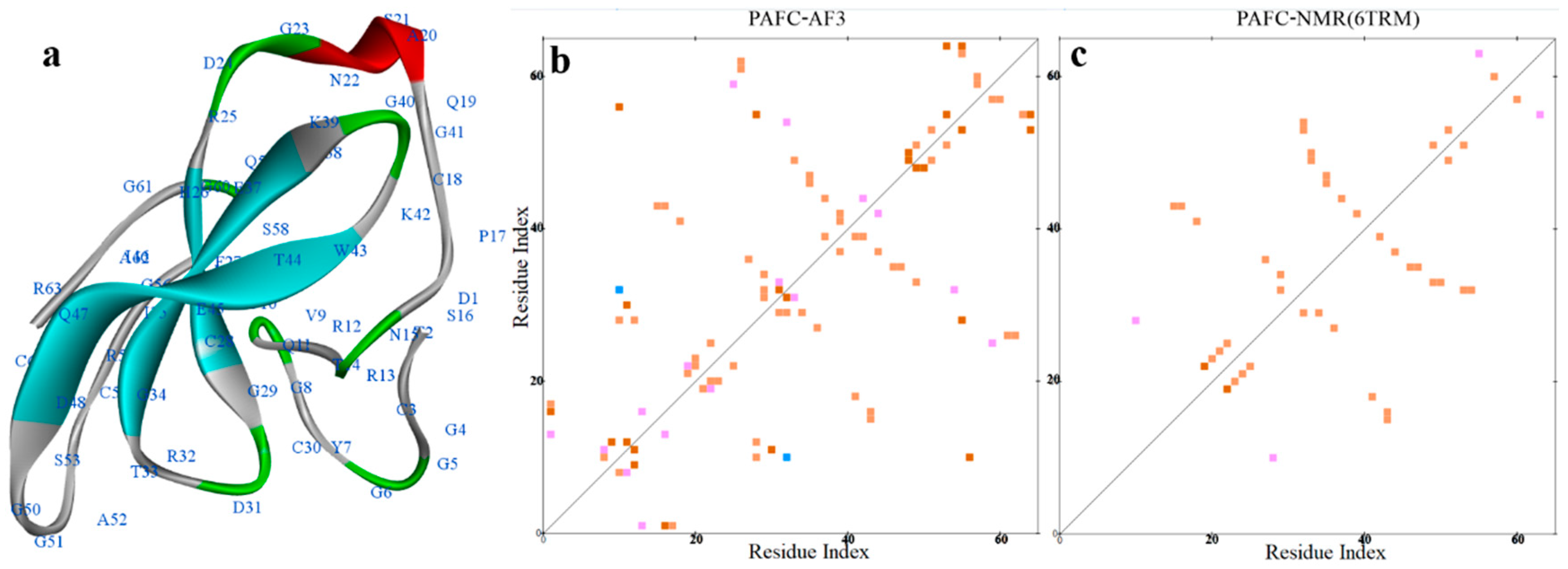
2.5. Structural Analysis of Other AFPs
3. Discussion
Supplementary Materials
Author Contributions
Acknowledgements
References
- Ashish Vaswani, N., Niki Parmar, Jakob Uszkoreit, Llion Jones, Aidan N. Gomez, Łukasz Kaiser., Attention Is All You Need. Arxiv, 2017.
- Buchan, D. W. A.; Jones, D. T. , Improved protein contact predictions with the MetaPSICOV2 server in CASP12. Proteins 2018, 86, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikov, S.; Park, H.; Kim, D. E.; Liu, Y. A.; Wang, R. Y. R.; Baker, D. , Structure prediction using sparse simulated NOE restraints with Rosetta in CASP11. Proteins 2016, 84, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Burley, S. K.; Berman, H. M.; Bhikadiya, C.; Bi, C. X.; Chen, L.; Di Costanzo, L.; et al. Protein Data Bank, J., Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019; 47, D520–D528. [Google Scholar]
- Godzik, A. , Metagenomics and the protein universe. Curr. Opin. Struct. Biol. 2011, 21, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Jones, D. T.; Thornton, J. M. , The impact of AlphaFold2 one year on. Nat. Methods 2022, 19, 15–20. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; et al. Hassabis, D., Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.; Simpkin, A. J.; Hartmann, M. D.; Rigden, D. J.; Keegan, R. M.; Lupas, A. N. , High-accuracy protein structure prediction in CASP14. Proteins 2021, 89, 1687–1699. [Google Scholar] [CrossRef] [PubMed]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Zídek, A.; et al. Hassabis, D., Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 59–596. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y. J.; Zhang, N.; Bersch, B.; Fidelis, K.; Inouye, M.; Ishida, Y.; et al. Montelione, G. T., Assessment of prediction methods for protein structures determined by NMR in CASP14: Impact of AlphaFold2. Proteins 2021, 89, 1959–1976. [Google Scholar] [CrossRef] [PubMed]
- Fowler, N. J.; Williamson, M. P. , The accuracy of protein structures in solution determined by AlphaFold and NMR. Structure 2022, 30, 925–933. [Google Scholar] [CrossRef]
- Fizil, A.; Gaspári, Z.; Barna, T.; Marx, F.; Batta, G. , "Invisible" Conformers of an Antifungal Disulfide Protein Revealed by Constrained Cold and Heat Unfolding, CEST-NMR Experiments, and Molecular Dynamics Calculations. Chem.-Eur. J. 2015, 21, 5136–5144. [Google Scholar] [CrossRef]
- Marx, F.; Binder, U.; Leiter, E.; Pocsi, I. , The Penicillium chrysogenum antifungal protein PAF, a promising tool for the development of new antifungal therapies and fungal cell biology studies. Cell Mol Life Sci 2008, 65, 445–54. [Google Scholar] [CrossRef] [PubMed]
- Sonderegger, C.; Fizil, A.; Burtscher, L.; Hajdu, D.; Muñoz, A.; Gáspári, Z.; et al. Marx, F., D19S Mutation of the Cationic, Cysteine-Rich Protein PAF: Novel Insights into Its Structural Dynamics, Thermal Unfolding and Antifungal Function. PLoS One 2017, 12, 21. [Google Scholar] [CrossRef]
- Batta, G.; Barna, T.; Gaspari, Z.; Sandor, S.; Kover, K. E.; Binder, U.; et al. Marx, F., Functional aspects of the solution structure and dynamics of PAF--a highly-stable antifungal protein from Penicillium chrysogenum. FEBS J 2009, 276, 2875–90. [Google Scholar] [CrossRef] [PubMed]
- Váradi, G.; Batta, G.; Galgóczy, L.; Hajdu, D.; Fizil, A.; Czajlik, A.; et al. Tóth, G. K., Confirmation of the Disulfide Connectivity and Strategies for Chemical Synthesis of the Four-Disulfide-Bond-Stabilized Aspergillus giganteus Antifungal Protein, AFP. J. Nat. Prod. 2023, 9. [Google Scholar]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; et al. Jumper, J. M., Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 24. [Google Scholar] [CrossRef]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; et al. Hassabis, D., Protein complex prediction with AlphaFold-Multimer. bioRxiv, 2021; 2021.10.04.463034. [Google Scholar]
- Židek, A. , AlphaFold v.2.3.0 Technical Note. GitHub https://github.com/google-deepmind/ aalphafold/blob/main/docs/technical_note_v2.3.0.md (2022).
- Karras, T.; Aittala, M.; Aila, T.; Laine, S. Elucidating the Design Space of Diffusion-Based Generative Models, 36th Conference on Neural Information Processing Systems (NeurIPS), Electr Network, Nov 28-Dec 09, 2022; Neural Information Processing Systems (Nips): Electr Network, 2022. [Google Scholar]
- Watson, E. R.; Novick, S.; Matyskiela, M. E.; Chamberlain, P. P.; de la Pena, A. H.; Zhu, J. Y.; et al. Lander, G. C., Molecular glue CELMoD compounds are regulators of cereblon conformation. Science 2022, 378, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Phukhamsakda, C.; Nilsson, R. H.; Bhunjun, C. S.; de Farias, A. R. G.; Sun, Y. R.; Wijesinghe, S. N.; et al. Hyde, K. D., The numbers of fungi: contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Divers. 2022, 114, 327–386. [Google Scholar] [CrossRef]
- Fisher, M. C.; Alastruey-Izquierdo, A.; Berman, J.; Bicanic, T.; Bignell, E. M.; Bowyer, P.; et al. Verweij, P. E., Tackling the emerging threat of antifungal resistance to human health. Nat. Rev. Microbiol. 2022, 20, 557–571. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M. C.; Gurr, S. J.; Cuomo, C. A.; Blehert, D. S.; Jin, H. L.; Stukenbrock, E. H.; et al. Cowen, L. E., Threats Posed by the Fungal Kingdom to Humans, Wildlife, and Agriculture. mBio 2020, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yamdeu, J. H. G.; Gong, Y. Y.; Orfila, C. , A review of postharvest approaches to reduce fungal and mycotoxin contamination of foods. Compr. Rev. Food. Sci. Food Saf. 2020, 19, 1521–1560. [Google Scholar] [CrossRef]
- Berman, J.; Krysan, D. J. , Drug resistance and tolerance in fungi. Nat. Rev. Microbiol. 2020, 18, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Denning, D. W. , Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. 2024, 24, e428–e438. [Google Scholar] [CrossRef] [PubMed]
- Kainz, K.; Bauer, M. A.; Madeo, F.; Carmona-Gutierrez, D. , Fungal infections in humans: the silent crisis. Microb. Cell 2020, 7, 143–145. [Google Scholar] [CrossRef]
- Galgóczy, L.; Yap, A.; Marx, F. , Cysteine-Rich Antifungal Proteins from Filamentous Fungi are Promising Bioactive Natural Compounds in Anti-Candida Therapy. Isr. J. Chem. 2019, 59, 360–370. [Google Scholar] [CrossRef]
- Palicz, Z.; Gáll, T.; Leiter, E.; Kollár, S.; Kovács, I.; Miszti-Blasius, K.; et al. Szentesi, P., Application of a low molecular weight antifungal protein from Penicillium chrysogenum (PAF) to treat pulmonary aspergillosis in mice. Emerg. Microbes Infect. 2016, 5, 9. [Google Scholar] [CrossRef]
- Palicz, Z.; Jenes, A.; Gáll, T.; Miszti-Blasius, K.; Kollár, S.; Kovács, I.; et al. Szentesi, P., In vivo application of a small molecular weight antifungal protein of Penicillium chrysogenum (PAF). Toxicol. Appl. Pharmacol. 2013, 269, 8–16. [Google Scholar] [CrossRef]
- Szappanos, H.; Szigeti, G. P.; Pal, B.; Rusznak, Z.; Szucs, G.; Rajnavolgyi, E.; et al. Csernoch, L., The Penicillium chrysogenum-derived antifungal peptide shows no toxic effects on mammalian cells in the intended therapeutic concentration. Naunyn Schmiedebergs Arch Pharmacol 2005, 371, 122–32. [Google Scholar] [CrossRef] [PubMed]
- Barna, B.; Leiter, E.; Hegedus, N.; Bíro, T.; Pócsi, I. , Effect of the Penicillium chrysogenum antifungal protein (PAF) on barley powdery mildew and wheat leaf rust pathogens. J. Basic Microbiol. 2008, 48, 516–520. [Google Scholar] [CrossRef]
- Holzknecht, J.; Marx, F. , Navigating the fungal battlefield: cysteine-rich antifungal proteins and peptides from Eurotiales. Front Fungal Biol 2024, 5, 1451455. [Google Scholar] [CrossRef]
- Huber, A.; Hajdu, D.; Bratschun-Khan, D.; Gaspari, Z.; Varbanov, M.; Philippot, S.; et al. Batta, G., New Antimicrobial Potential and Structural Properties of PAFB: A Cationic, Cysteine-Rich Protein from Penicillium chrysogenum Q176. Sci Rep 2018, 8, 1751. [Google Scholar] [CrossRef]
- Camposolivas, R.; Bruix, M.; Santoro, J.; Lacadena, J.; Delpozo, A. M.; Gavilanes, J. G.; Rico, M. , Nmr Solution Structure of the Antifungal Protein from Aspergillus-Giganteus - Evidence For Cysteine Pairing Isomerism. Biochemistry 1995, 34, 3009–3021. [Google Scholar] [CrossRef]
- Galgóczy, L.; Borics, A.; Virágh, M.; Ficze, H.; Váradi, G.; Kele, Z.; Marx, F. , Structural determinants of Neosartorya fischeri antifungal protein (NFAP) for folding, stability and antifungal activity. Scientific Reports 2017, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z. T.; Ao, J. Q.; Yang, W. C.; Jiao, L. P.; Zheng, T. L.; Chen, X. H. , Purification and characterization of a novel antifungal protein secreted by Penicillium chrysogenum from an Arctic sediment. Appl. Microbiol. Biotechnol. 2013, 97, 10381–10390. [Google Scholar] [CrossRef] [PubMed]
- Toth, L.; Kele, Z.; Borics, A.; Nagy, L. G.; Varadi, G.; Viragh, M.; et al. Galgoczy, L., NFAP2, a novel cysteine-rich anti-yeast protein from Neosartorya fischeri NRRL 181: isolation and characterization. AMB Express 2016, 6, 75. [Google Scholar] [CrossRef]
- Virágh, M.; Marton, A.; Vizler, C.; Tóth, L.; Vágvölgyi, C.; Marx, F.; Galgóczy, L. , Insight into the antifungal mechanism of Neosartorya fischeri antifungal protein. Protein Cell 2015, 6, 518–528. [Google Scholar] [CrossRef]
- Kaiserer, L.; Oberparleiter, C.; Weiler-Görz, R.; Burgstaller, W.; Leiter, E.; Marx, F. , Characterization of the Penicillium chrysogenum antifungal protein PAF. Arch. Microbiol. 2003, 180, 204–210. [Google Scholar] [CrossRef]
- Virágh, M.; Vörös, D.; Kele, Z.; Kovács, L.; Fizil, A.; Lakatos, G.; et al. Galgóczy, L., Production of a defensin-like antifungal protein NFAP from Neosartorya fischeri in Pichia pastoris and its antifungal activity against filamentous fungal isolates from human infections. Protein Expr. Purif. 2014, 94, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V. , A small protein that fights fungi: AFP as a new promising antifungal agent of biotechnological value. Appl. Microbiol. Biotechnol. 2008, 78, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Huber, A.; Oemer, G.; Malanovic, N.; Lohner, K.; Kovács, L.; Salvenmoser, W.; et al. Marx, F., Membrane Sphingolipids Regulate the Fitness and Antifungal Protein Susceptibility of Neurospora crassa. Front. Microbiol. 2019, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Huber, A.; Galgoczy, L.; Varadi, G.; Holzknecht, J.; Kakar, A.; Malanovic, N.; et al. Marx, F., Two small, cysteine-rich and cationic antifungal proteins from Penicillium chrysogenum: A comparative study of PAF and PAFB. Biochim Biophys Acta Biomembr 2020, 1862, 183246. [Google Scholar] [CrossRef] [PubMed]
- Holzknecht, J.; Kuhbacher, A.; Papp, C.; Farkas, A.; Varadi, G.; Marcos, J. F.; et al. Marx, F., The Penicillium chrysogenum Q176 Antimicrobial Protein PAFC Effectively Inhibits the Growth of the Opportunistic Human Pathogen Candida albicans. J Fungi (Basel) 2020, 6. [Google Scholar]
- Hegedus, N.; Leiter, É.; Kovács, B.; Tomori, V.; Kwon, N. J.; Emri, T.; et al. Pócsi, I., The small molecular mass antifungal protein of Penicillium chrysogenum - a mechanism of action oriented review. J. Basic Microbiol. 2011, 51, 561–571. [Google Scholar] [CrossRef]
- Moreno, A. B.; del Pozo, A. M.; Segundo, B. S. , Biotechnologically relevant enzymes and proteins -: Antifungal mechanism of the Aspergillus giganteusAFP against the rice blast fungus Magnaporthe grisea. Appl. Microbiol. Biotechnol. 2006, 72, 883–895. [Google Scholar] [CrossRef] [PubMed]
- Kovács, R.; Nagy, F.; Tóth, Z.; Forgács, L.; Tóth, L.; Váradi, G.; et al. Galgóczy, L., The Neosartorya fischeri Antifungal Protein 2 (NFAP2): A New Potential Weapon against Multidrug-Resistant Candida auris Biofilms. Int. J. Mol. Sci. 2021, 22, 14. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martín, A.; Acosta, R.; Liddell, S.; Núñez, F.; Benito, M. J.; Asensio, M. A. , Characterization of the novel antifungal protein PgAFP and the encoding gene of Penicillium chrysogenum. Peptides 2010, 31, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Czajlik, A.; Holzknecht, J.; Galgoczy, L.; Toth, L.; Poor, P.; Ordog, A.; et al. Batta, G., Solution Structure, Dynamics, and New Antifungal Aspects of the Cysteine-Rich Miniprotein PAFC. Int J Mol Sci 2021, 22. [Google Scholar]
- Kovács, R.; Holzknecht, J.; Hargital, Z.; Papp, C.; Farkas, A.; Borics, A.; et al. Galgóczy, L., In Vivo Applicability of Neosartorya fischeri Antifungal Protein 2 (NFAP2) in Treatment of Vulvovaginal Candidiasis. Antimicrob. Agents Chemother. 2019, 63, 12. [Google Scholar] [CrossRef] [PubMed]
- Oberparleiter, C.; Kaiserer, L.; Haas, H.; Ladurner, P.; Andratsch, M.; Marx, F. , Active internalization of the Penicillium chrysogenum antifungal protein PAF in sensitive aspergilli. Antimicrob. Agents Chemother. 2003, 47, 3598–3601. [Google Scholar] [CrossRef] [PubMed]
- Hajdu, D.; Huber, A.; Czajlik, A.; Tóth, L.; Kele, Z.; Kocsubé, S.; et al. Batta, G., Solution structure and novel insights into phylogeny and mode of action of the Neosartorya (Aspergillus) fischeri antifungal protein (NFAP). Int. J. Biol. Macromol. 2019, 129, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Czajlik, A.; Batta, A.; Kerner, K.; Fizil, A.; Hajdu, D.; Raics, M.; et al. Batta, G., DMSO-Induced Unfolding of the Antifungal Disulfide Protein PAF and Its Inactive Variant: A Combined NMR and DSC Study. Int J Mol Sci 2023, 24. [Google Scholar]
- Tóth, L.; Váradi, G.; Borics, A.; Batta, G.; Kele, Z.; Vendrinszky, A.; et al. Galgóczy, L., Anti-Candidal Activity and Functional Mapping of Recombinant and Synthetic Neosartorya fischeri Antifungal Protein 2 (NFAP2). Front. Microbiol. 2018, 9, 12. [Google Scholar] [CrossRef]
- Váradi, G.; Kele, Z.; Czajlik, A.; Borics, A.; Bende, G.; Papp, C.; et al. Galgóczy, L., Hard nut to crack: Solving the disulfide linkage pattern of the Neosartorya (Aspergillus) fischeri antifungal protein 2. Protein Sci. 2023, 32, 13. [Google Scholar] [CrossRef]
- Pavela, O.; Juhász, T.; Tóth, L.; Czajlik, A.; Batta, G.; Galgóczy, L.; Beke-Somfai, T. , Mapping of the Lipid-Binding Regions of the Antifungal Protein NFAP2 by Exploiting Model Membranes. J. Chem Inf. Model. 2024, 64, 6557–6569. [Google Scholar] [CrossRef] [PubMed]
- Sonderegger, C.; Varadi, G.; Galgoczy, L.; Kocsube, S.; Posch, W.; Borics, A.; et al. Marx, F., The Evolutionary Conserved gamma-Core Motif Influences the Anti-Candida Activity of the Penicillium chrysogenum Antifungal Protein PAF. Front Microbiol 2018, 9, 1655. [Google Scholar] [CrossRef] [PubMed]
- Chen, C. J.; Wu, Y.; Li, J. W.; Wang, X.; Zeng, Z. H.; Xu, J.; et al. Xia, R., TBtools-II: A "one for all, all for one"bioinformatics platform for biological big-data mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Alexander, L. T.; Lepore, R.; Kryshtafovych, A.; Adamopoulos, A.; Alahuhta, M.; Arvin, A. M.; et al. Schwede, T., Target highlights in CASP14: Analysis of models by structure providers. Proteins 2021, 89, 1647–1672. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R. F. , A Simple Method for Displaying the Hydropathic Character of A Protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Fizil, A.; Sonderegger, C.; Czajlik, A.; Fekete, A.; Komaromi, I.; Hajdu, D.; et al. Batta, G., Calcium binding of the antifungal protein PAF: Structure, dynamics and function aspects by NMR and MD simulations. PLoS One 2018, 13, e0204825. [Google Scholar] [CrossRef] [PubMed]
- Utesch, T.; Catalina, A. D.; Schattenberg, C.; Paege, N.; Schmieder, P.; Krause, E.; et al. Mroginski, M. A., A Computational Modeling Approach Predicts Interaction of the Antifungal Protein AFP from Aspergillus giganteus with Fungal Membranes via Its γ-Core Motif. mSphere 2018, 3, 15. [Google Scholar] [CrossRef] [PubMed]
- Yount, N. Y.; Yeaman, M. R. , Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 7363–7368. [Google Scholar] [CrossRef]
- Yount, N. Y.; Yeaman, M. R. Emerging Themes and Therapeutic Prospects for Anti-Infective Peptides. In Annual Review of Pharmacology and Toxicology, Vol 52; Insel, P. A., Amara, S. G., Blaschke, T. F., Eds.; Annual Reviews: Palo Alto, 2012; Vol. 52, pp 337-+. [Google Scholar]
- Tejero, R.; Huang, Y. J.; Ramelot, T. A.; Montelione, G. T. , AlphaFold Models of Small Proteins Rival the Accuracy of Solution NMR Structures. Front. Mol. Biosci. 2022, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Mello, E.; Taveira, G. B.; de Oliveira Carvalho, A.; Gomes, V. M. , Improved smallest peptides based on positive charge increase of the gamma-core motif from PnuD(1) and their mechanism of action against Candida species. Int J Nanomedicine 2019, 14, 407–420. [Google Scholar] [CrossRef] [PubMed]
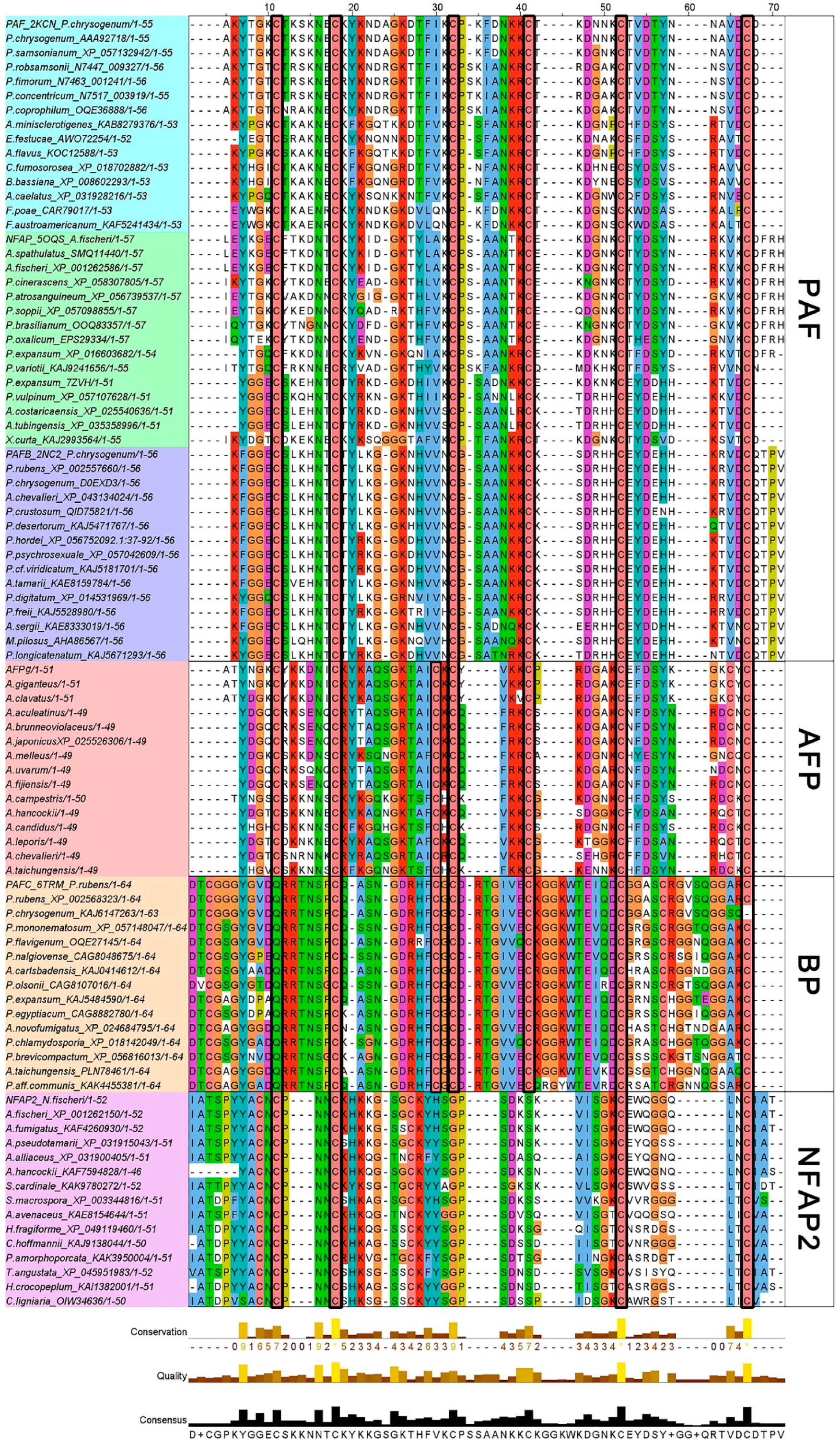
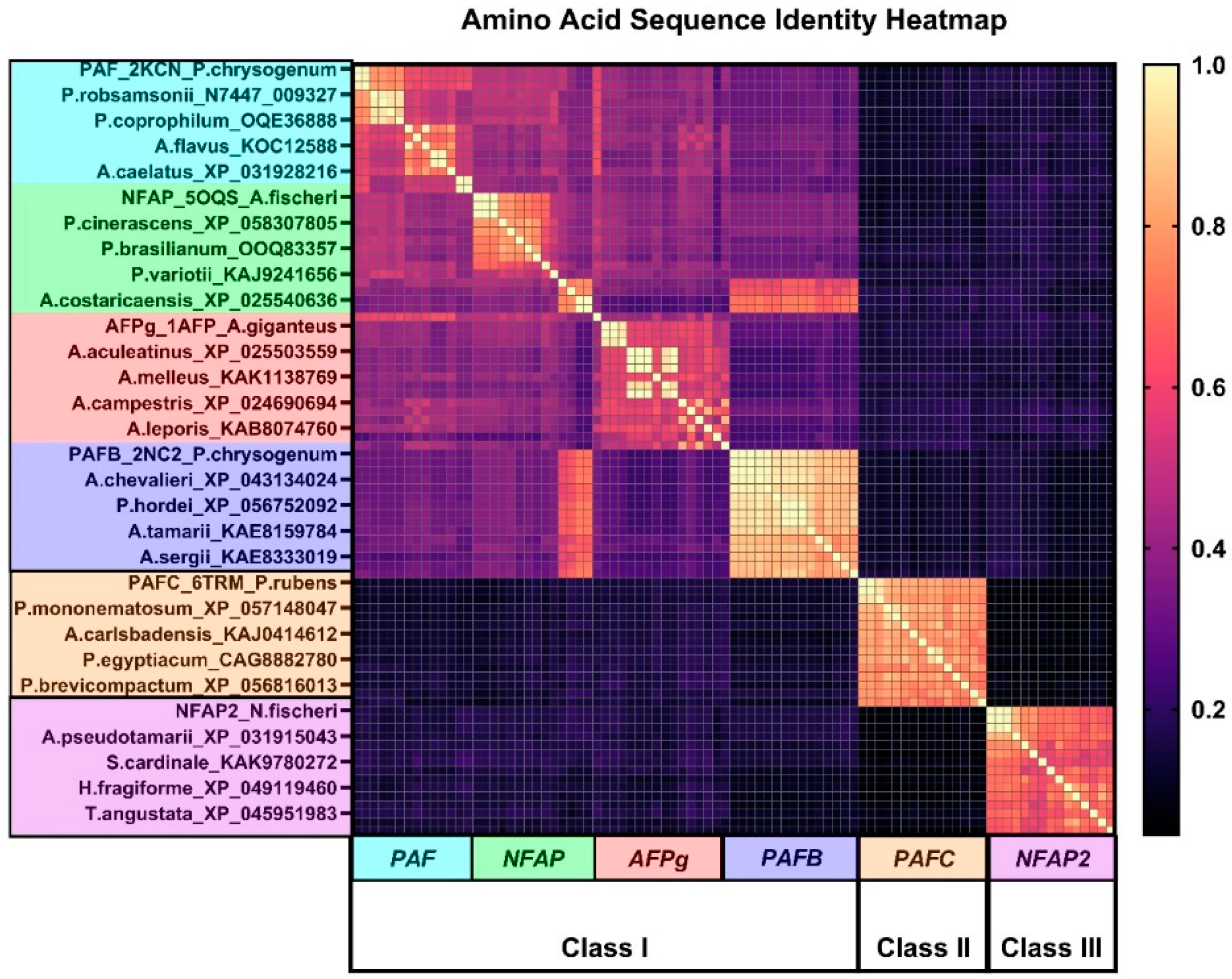
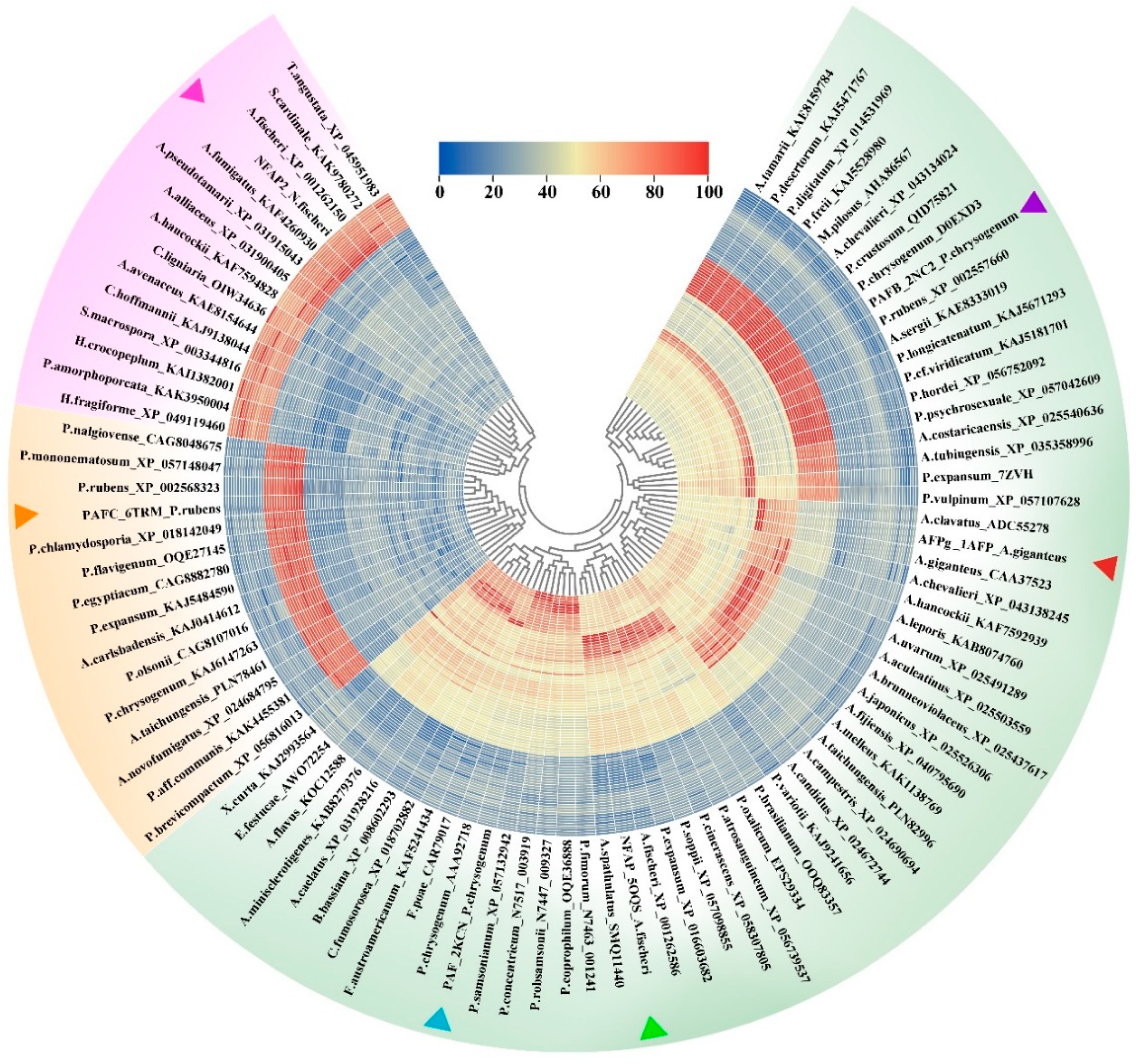
| PAF | NFAP | AFPg | PAFB | PAFC | NFAP2 | |
|---|---|---|---|---|---|---|
| Organism |
Penicillium chrysogenum Q176 |
Neosartorya (Aspergillus) fischeri NRRL 181 |
Aspergillus giganteus |
Penicillium chrysogenum Q176 |
Penicillium chrysogenum Q176 |
Neosartorya (Aspergillus) fischeri NRRL 181 |
| NMR | 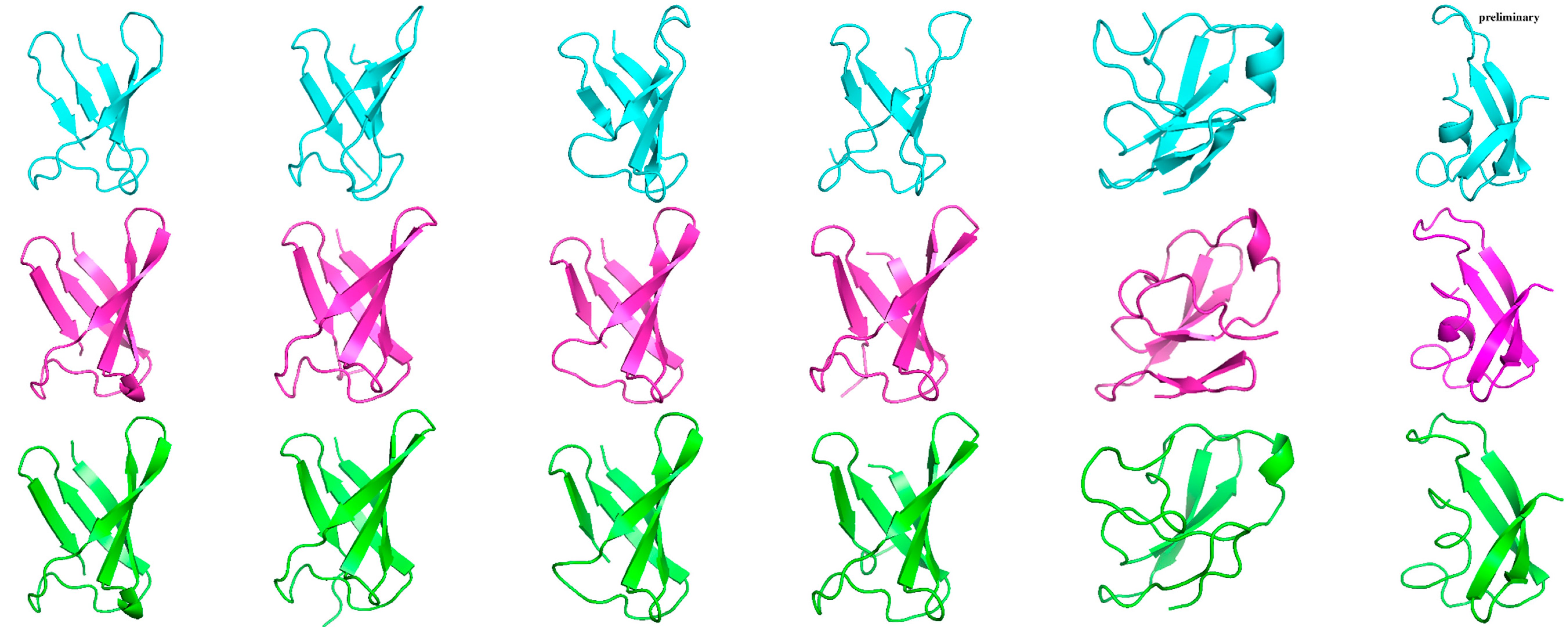 |
|||||
| AF2 | ||||||
| AF3 | ||||||
| RMSD to NMR structure | AF2(1.497); AF3(1.429) | AF2(1.429); AF3(1.395) | AF2(1.004); AF3(0.865) | AF2(1.795); AF3(1.648) | AF2(3.129); AF3(0.868) |
AF2(1.244); AF3(1.257) |
| Disulfide bond pattern |
abcabc: 7-36, 14-43, 28-54 |
abcabc: 7-35, 14-42, 27-53 |
abcdabcd: 26-49, 28-51 (NMR); 7-33, 14-40, 26-49, 28-51 (AF3) |
abcabc: 6-34, 13-41, 26-52 |
abcabdcd: 3-30, 18-38, 28-54, 49-64 |
abbcac: 9-40, 11-15, 23-49 |
| Correctness of disulfide bonds | AF2(Y); AF3(Y) | AF2(N); AF3(Y) | AF2(N); AF3(?) | AF2(N); AF3(Y) | AF2(N); AF3(Y) | AF2(Y); AF3(Y) |
| Ions Num | Mg2+ | 2Mg2+ | 3Mg2+ | 4Mg2+ | ||||
|---|---|---|---|---|---|---|---|---|
| Score | ipTM | pTM | ipTM | pTM | ipTM | pTM | ipTM | pTM |
| PAF | 0.78 | 0.85 | 0.7 | 0.85 | 0.58 | 0.84 | 0.55 | 0.84 |
| PAFD19S | 0.79 | 0.85 | 0.71 | 0.85 | 0.57 | 0.84 | 0.53 | 0.84 |
| Ions Num | Na+ | 2Na+ | 3Na+ | 4Na+ | ||||
| Score | ipTM | pTM | ipTM | pTM | ipTM | pTM | ipTM | pTM |
| PAF | 0.8 | 0.85 | 0.75 | 0.86 | 0.69 | 0.85 | 0.65 | 0.85 |
| PAFD19S | 0.8 | 0.85 | 0.75 | 0.85 | 0.69 | 0.85 | 0.65 | 0.85 |
| Ions | Ca2+ | 2Ca2+ | 3Ca2+ | 4Ca2+ | ||||
| Score | ipTM | pTM | ipTM | pTM | ipTM | pTM | ipTM | pTM |
| PAF | 0.74 | 0.84 | 0.65 | 0.85 | 0.61 | 0.84 | 0.57 | 0.84 |
| PAFD19S | 0.77 | 0.84 | 0.69 | 0.85 | 0.66 | 0.85 | 0.55 | 0.84 |
| Ions | Ca2+andMg2+ | Ca2+andNa+ | Na+andMg2+ | |||||
| Score | ipTM | pTM | ipTM | pTM | ipTM | pTM | ||
| PAF | 0.78 | 0.85 | 0.78 | 0.85 | 0.81 | 0.86 | ||
| PAFD19S | 0.78 | 0.85 | 0.78 | 0.85 | 0.81 | 0.86 | ||
| Metric | NMR- PAF |
NMR- NFAP |
NMR- PAFB |
NMR- PAFC |
NMR- AFPg |
AF2- PAF |
AF2- NFAP |
AF2- PAFB |
AF2- PAFC |
AF2- AFPg |
AF2- NFAP2 |
AF3- PAF |
AF3- NFAP |
AF3- PAFB |
AF3- PAFC |
AF3- AFPg |
AF3- NFAP2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MolProbity Score | 1.98 | 3.19 | 3.52 | 2.58 | 4.44 | 1.86 | 2.62 | 2.99 | 3.49 | 2.18 | 1.75 | 0.673 | 1.70 | 1.32 | 0.99 | 1.31 | 1.31 |
| Clashscore | 1.29 | 8.84 | 17.06 | 3.46 | 89.22 | 24.11 | 33.38 | 41.82 | 47.21 | 39.79 | 11.01 | 1.563 | 13.62 | 5.88 | 2.31 | 4.59 | 3.53 |
| Poor rotamers(%) | 35.3 | 33.43 | 34.18 | 21.80 | 36.34 | 0 | 4.57 | 3.4 | 5.67 | 0.78 | 2.33 | 0 | 0.65 | 0 | 0 | 0.78 | 0.78 |
| Favored rotamers(%) | 43.2 | 45.39 | 40.31 | 63.72 | 37.03 | 100 | 92.16 | 92.52 | 88.65 | 99.22 | 93.80 | 100 | 99.35 | 99.32 | 100 | 98.45 | 99.22 |
|
Ramachandran Outliers(%) |
1.796 | 6.36 | 2.78 | 1.61 | 10.87 | 0 | 0 | 5.56 | 21.50 | 1.36 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
Ramachandran favored(%) |
97.92 | 89.18 | 86.23 | 92.83 | 68.52 | 99.73 | 96.97 | 90.12 | 71.51 | 97,96 | 98 | 99.3 | 99.39 | 100 | 100 | 100 | 98.67 |
| Rama distribution Z-score |
-1.88 ±1.04 |
-3.36 ±0.82 |
-3.57 ±0.95 |
-1.70 ±0.79 |
-5.86 ±1.01 |
0.72 ±0.95 |
0.34± 1.03 |
-1.11 ±1.04 |
-4.62 ±0.70 |
-0.85 ±1.06 |
-1.35 ±1.02 |
1.32 ±1.03 |
0.89 ±1.03 |
1.14 ±1.04 |
0.56 ±1.05 |
0.35 ±1.22 |
-0.73 ±1.1 |
| Cβ deviations >0.25Å(%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Bad bonds(%) | 0 | 0 | 0 | 0 | 0 | 2.82 | 2.92 | 2.47 | 4.92 | 3.11 | 3.64 | 0 | 0.14 | 0 | 0 | 0 | 0 |
| Bad angles(%) | 0 | 0 | 0 | 0 | 0 | 0.8 | 0.91 | 1.46 | 4.28 | 0.75 | 0.88 | 0 | 0.16 | 00 | 0 | 0 | 0 |
| Cis Prolines (Per Chain) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 1/3 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 1/3 |
| CaBLAM outliers(%) | 3.81 | 4.09 | 3.80 | 2.86 | 13.83 | 0 | 0 | 1.90 | 6.1 | 0.7 | 1.4 | 0 | 0 | 0 | 2.23 | 0 | 2.77 |
| CA Geometry outliers(%) | 1.96 | 0 | 0.10 | 1.67 | 1.60 | 0 | 0 | 0.64 | 2.22 | 0 | 4.17 | 0 | 0 | 0 | 0 | 0 | 3.47 |
| Chiral volume outliers | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Waters with clashes(%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





