Submitted:
30 March 2024
Posted:
02 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
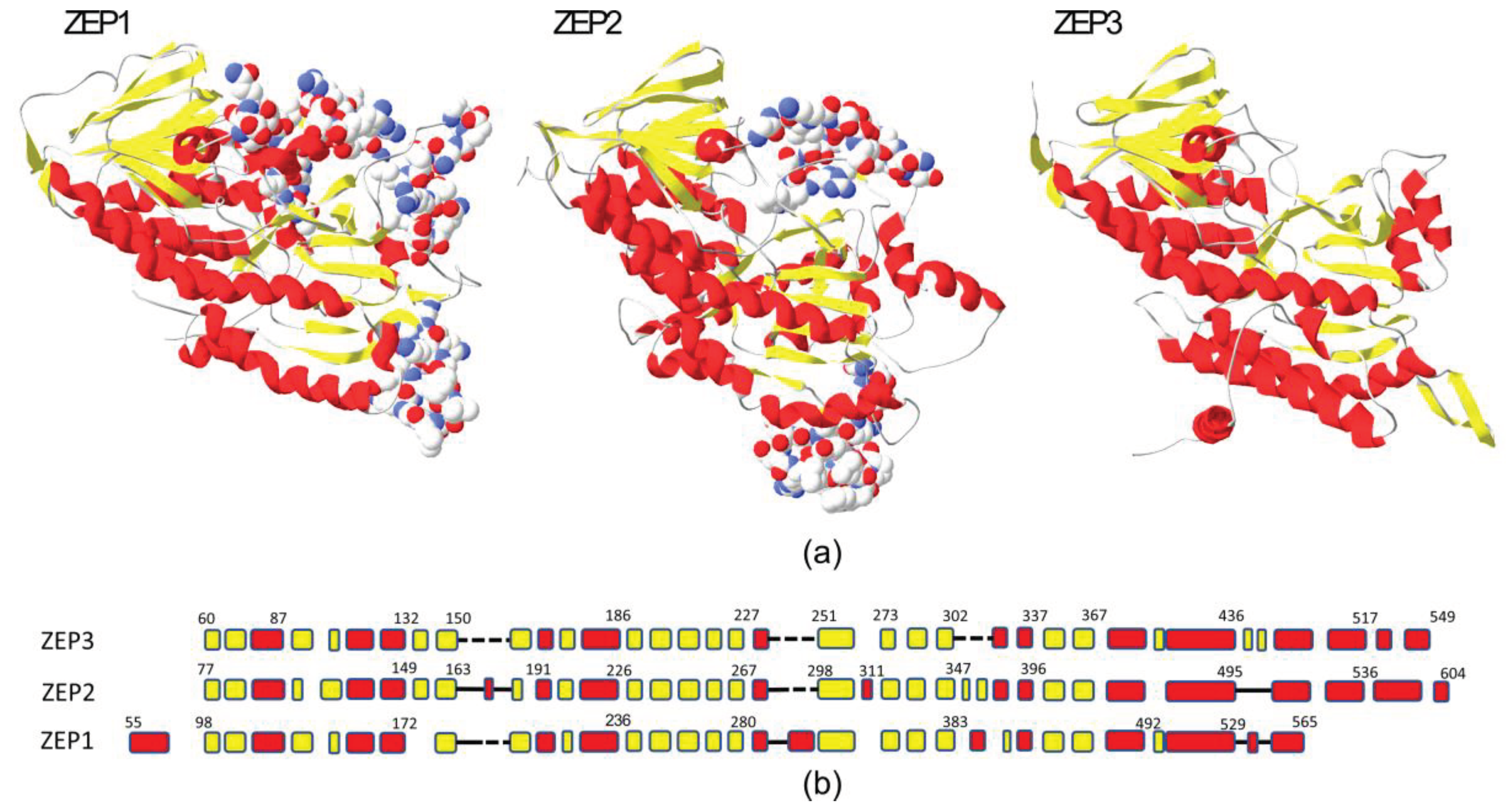
2.1. Phylogenetic and Structural Study of ZEP Genes
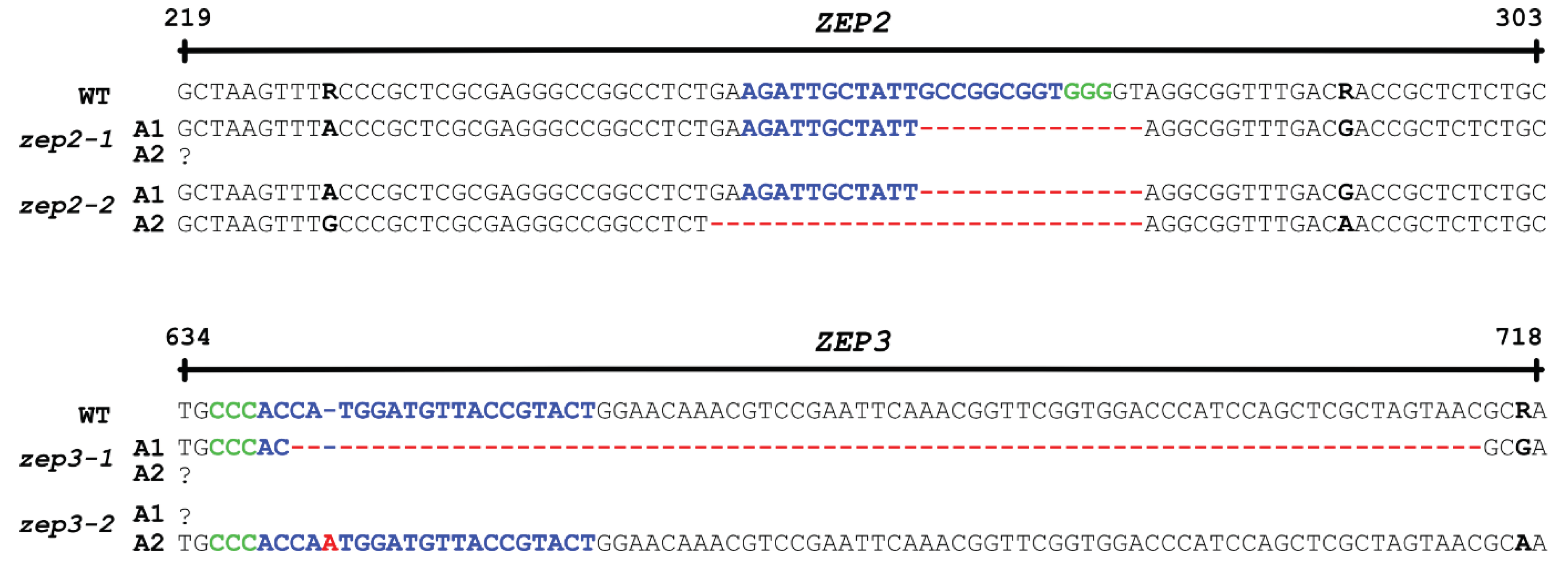
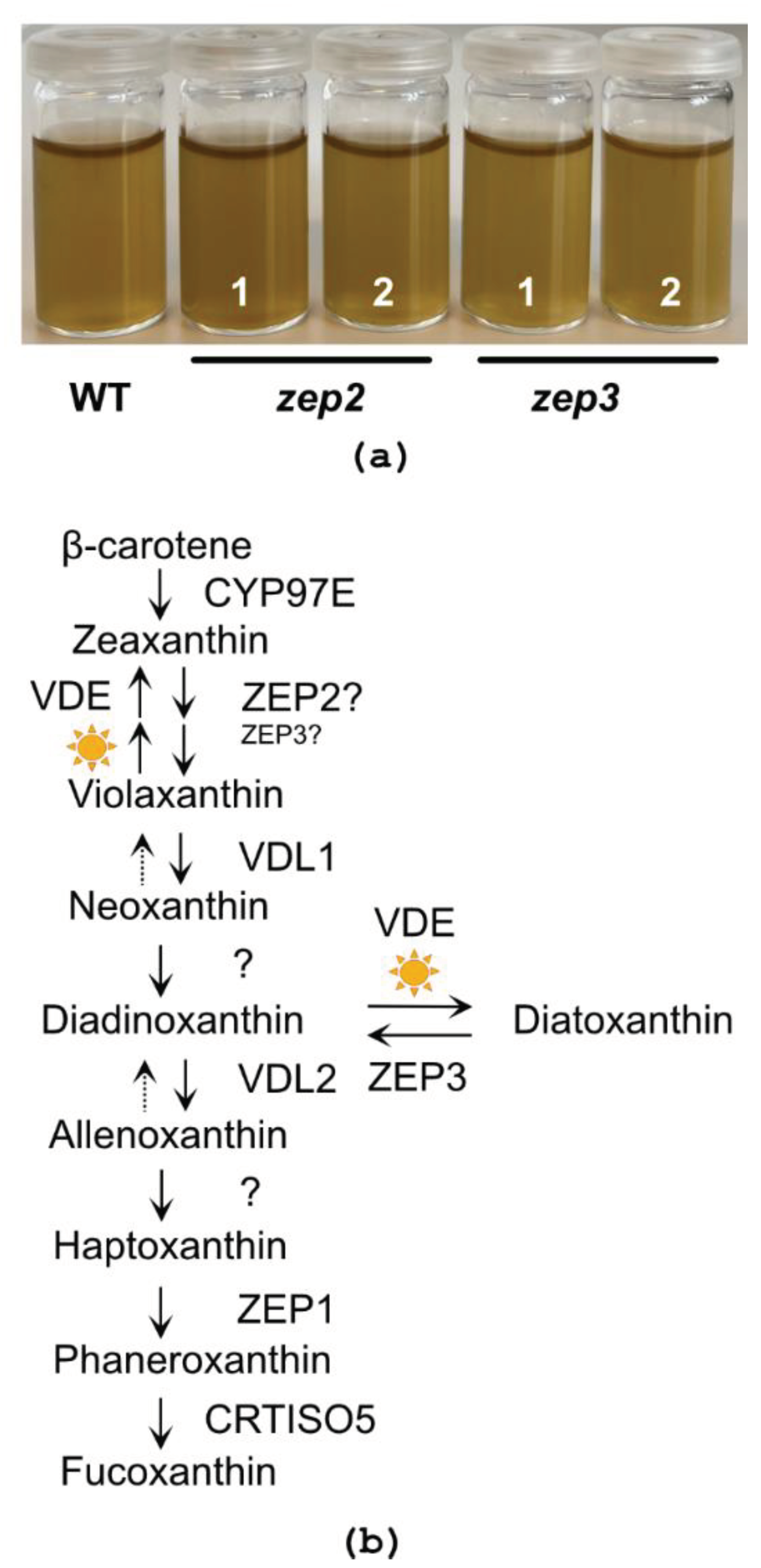
2.2. CRISPR/Cas9-Generated zep2 and zep3 Knockout Mutants
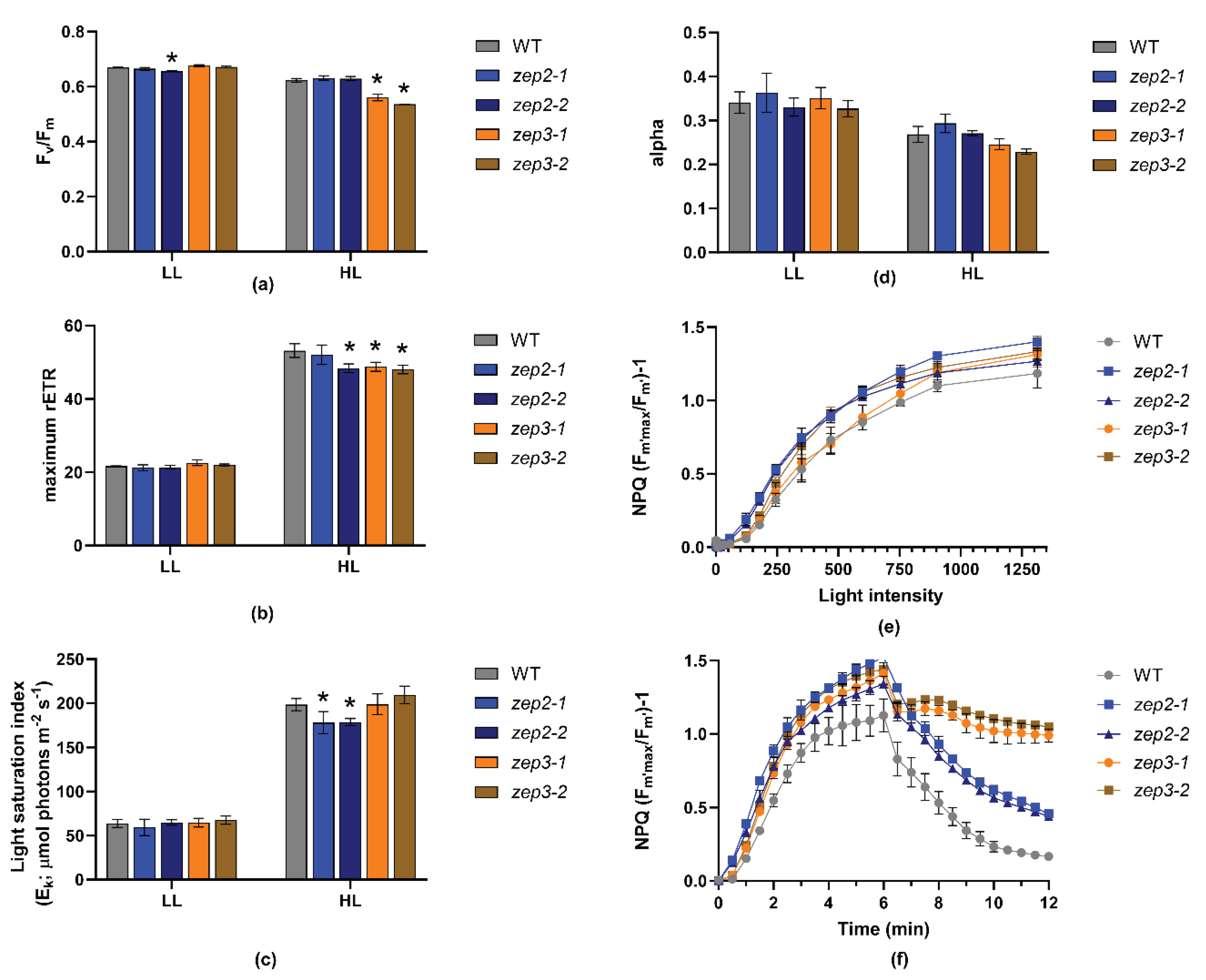
2.3. Loss of ZEP3 Blocks the Back-Conversion of Diatoxanthin to Diadinoxanthin in Low Light
2.4. Loss of ZEP3 Inhibits Relaxation of the Photoprotective Mechanism NPQ
3. Materials and Methods
3.1. Structural Comparison and Phylogenetic Analyses of ZEP Proteins
3.2. CRISPR/Cas9 Gene Editing of the ZEP2 and ZEP3 Genes
3.3. Light Conditions
3.4. Growth Rates
3.5. Measurements of Photosynthetic Parameters
3.6. Diatoxanthin In Vivo Stability Experiment (Pigment Analyses)
3.7. Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sandmann, G. Diversity and Origin of Carotenoid Biosynthesis: Its History of Coevolution towards Plant Photosynthesis. New Phytol 2021, 232, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Meléndez-Martínez, A.J. An Overview of Carotenoids, Apocarotenoids, and Vitamin A in Agro-Food, Nutrition, Health, and Disease. Molecular Nutrition Food Res 2019, 63, 1801045. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Sun, H.; Deng, J.; Huang, J.; Chen, F. Carotenoid Production from Microalgae: Biosynthesis, Salinity Responses and Novel Biotechnologies. Mar Drugs 2021, 19, 713. [Google Scholar] [CrossRef] [PubMed]
- Kabir, Md.T.; Rahman, H.; Shah, M.; Jamiruddin, M.R.; Basak, D.; Al-Harrasi, A.; Bhatia, S.; Ashraf, G.M.; Najda, A.; El-kott, A.F.; et al. Therapeutic Promise of Carotenoids as Antioxidants and Anti-Inflammatory Agents in Neurodegenerative Disorders. Biomed Pharmacother 2022, 146, 112610. [Google Scholar] [CrossRef] [PubMed]
- Seth, K.; Kumar, A.; Rastogi, R.P.; Meena, M.; Vinayak, V. Harish Bioprospecting of Fucoxanthin from Diatoms — Challenges and Perspectives. Algal Res 2021, 60, 102475. [Google Scholar] [CrossRef]
- Pocha, C.K.R.; Chia, W.Y.; Chew, K.W.; Munawaroh, H.S.H.; Show, P.L. Current Advances in Recovery and Biorefinery of Fucoxanthin from Phaeodactylum Tricornutum. Algal Res 2022, 65, 102735. [Google Scholar] [CrossRef]
- Sathasivam, R.; Ki, J.-S. A Review of the Biological Activities of Microalgal Carotenoids and Their Potential Use in Healthcare and Cosmetic Industries. Mar Drugs 2018, 16, 26. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.; Pandey, V.K.; Dash, K.K.; Zanwar, S.; Singh, R. Natural Bio-Colorant and Pigments: Sources and Applications in Food Processing. J. Agric. Res. 2023, 12, 100628. [Google Scholar] [CrossRef]
- Ávila-Román, J.; García-Gil, S.; Rodríguez-Luna, A.; Motilva, V.; Talero, E. Anti-Inflammatory and Anticancer Effects of Microalgal Carotenoids. Mar Drugs 2021, 19, 531. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Yuan, J.-P.; Wu, C.-F.; Wang, J.-H. Fucoxanthin, a Marine Carotenoid Present in Brown Seaweeds and Diatoms: Metabolism and Bioactivities Relevant to Human Health. Mar Drugs 2011, 9, 1806–1828. [Google Scholar] [CrossRef] [PubMed]
- Galasso, C.; Corinaldesi, C.; Sansone, C. Carotenoids from Marine Organisms: Biological Functions and Industrial Applications. Antioxidants 2017, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Bae, M.; Kim, M.-B.; Park, Y.-K.; Lee, J.-Y. Health Benefits of Fucoxanthin in the Prevention of Chronic Diseases. Biochim Biophys Acta Mol Cell Biol Lipids. 2020, 1865, 158618. [Google Scholar] [CrossRef] [PubMed]
- Leong, Y.K.; Chen, C.-Y.; Varjani, S.; Chang, J.-S. Producing Fucoxanthin from Algae – Recent Advances in Cultivation Strategies and Downstream Processing. Bioresour. Technol. 2022, 344, 126170. [Google Scholar] [CrossRef] [PubMed]
- Nymark, M.; Valle, K.C.; Brembu, T.; Hancke, K.; Winge, P.; Andresen, K.; Johnsen, G.; Bones, A.M. An Integrated Analysis of Molecular Acclimation to High Light in the Marine Diatom Phaeodactylum Tricornutum. PLoS ONE 2009, 4, e7743. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.S. Photosynthetic Pigment Organization in Diatoms (Bacillariophyceae). J. Phycol. 1988, 24, 96–102. [Google Scholar] [CrossRef]
- Konishi, I.; Hosokawa, M.; Sashima, T.; Maoka, T.; Miyashita, K. Suppressive Effects of Alloxanthin and Diatoxanthin from Halocynthia Roretzi on LPS-Induced Expression of pro-Inflammatory Genes in RAW264.7 Cells. J. Oleo Sci. 2008, 57, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Pistelli, L.; Sansone, C.; Smerilli, A.; Festa, M.; Noonan, D.M.; Albini, A.; Brunet, C. MMP-9 and IL-1β as Targets for Diatoxanthin and Related Microalgal Pigments: Potential Chemopreventive and Photoprotective Agents. Mar. Drugs 2021, 19, 354. [Google Scholar] [CrossRef] [PubMed]
- Sansone, C.; Pistelli, L.; Del Mondo, A.; Calabrone, L.; Fontana, A.; Noonan, D.M.; Albini, A.; Brunet, C. The Microalgal Diatoxanthin Inflects the Cytokine Storm in SARS-CoV-2 Stimulated ACE2 Overexpressing Lung Cells. Antioxidants 2022, 11, 1515. [Google Scholar] [CrossRef] [PubMed]
- Sansone, C.; Pistelli, L.; Calabrone, L.; Del Mondo, A.; Fontana, A.; Festa, M.; Noonan, D.M.; Albini, A.; Brunet, C. The Carotenoid Diatoxanthin Modulates Inflammatory and Angiogenesis Pathways In Vitro in Prostate Cancer Cells. Antioxidants 2023, 12, 359. [Google Scholar] [CrossRef] [PubMed]
- Goss, R.; Lepetit, B. Biodiversity of NPQ. J Plant Physiol 2015, 172, 13–32. [Google Scholar] [CrossRef] [PubMed]
- Lavaud, J.; Goss, R. The Peculiar Features of Non-Photochemical Fluorescence Quenching in Diatoms and Brown Algae. In Non-photochemical quenching and energy dissipation in plants, algae and cyanobacteria. Advances in photosynthesis and respiration (Including bioenergy and related processes); Demmig-Adams, B., Garab, G., Adams, I.W., Govindjee, Eds.; Springer: Dordrecht, 2014; Vol. 40, pp. 421–443.
- Sharma, A.K.; Nymark, M.; Sparstad, T.; Bones, A.M.; Winge, P. Transgene-Free Genome Editing in Marine Algae by Bacterial Conjugation – Comparison with Biolistic CRISPR/Cas9 Transformation. Sci Rep 2018, 8, 14401. [Google Scholar] [CrossRef] [PubMed]
- Serif, M.; Dubois, G.; Finoux, A.L.; Teste, M.A.; Jallet, D.; Daboussi, F. One-Step Generation of Multiple Gene Knock-Outs in the Diatom Phaeodactylum Tricornutum by DNA-Free Genome Editing. Nat Commun 2018, 9, 3924. [Google Scholar] [CrossRef] [PubMed]
- Araújo, R.; Vázquez Calderón, F.; Sánchez López, J.; Azevedo, I.C.; Bruhn, A.; Fluch, S.; Garcia Tasende, M.; Ghaderiardakani, F.; Ilmjärv, T.; Laurans, M.; et al. Current Status of the Algae Production Industry in Europe: An Emerging Sector of the Blue Bioeconomy. Front. Mar. Sci. 2021, 7, 626389. [Google Scholar] [CrossRef]
- Dautermann, O.; Lyska, D.; Andersen-Ranberg, J.; Becker, M.; Fröhlich-Nowoisky, J.; Gartmann, H.; Krämer, L.C.; Mayr, K.; Pieper, D.; Rij, L.M.; et al. An Algal Enzyme Required for Biosynthesis of the Most Abundant Marine Carotenoids. Sci. Adv. 2020, 6, eaaw9183. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Cao, T.; Dautermann, O.; Buschbeck, P.; Cantrell, M.B.; Chen, Y.; Lein, C.D.; Shi, X.; Ware, M.A.; Yang, F.; et al. Green Diatom Mutants Reveal an Intricate Biosynthetic Pathway of Fucoxanthin. Proc. Natl. Acad. Sci. U.S.A. 2022, 119, e2203708119. [Google Scholar] [CrossRef]
- Cao, T.; Bai, Y.; Buschbeck, P.; Tan, Q.; Cantrell, M.B.; Chen, Y.; Jiang, Y.; Liu, R.-Z.; Ries, N.K.; Shi, X.; et al. An Unexpected Hydratase Synthesizes the Green Light-Absorbing Pigment Fucoxanthin. Plant Cell 2023, 35, 3053–3072. [Google Scholar] [CrossRef] [PubMed]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum Genome Reveals the Evolutionary History of Diatom Genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Coesel, S.; Obornik, M.; Varela, J.; Falciatore, A.; Bowler, C. Evolutionary Origins and Functions of the Carotenoid Biosynthetic Pathway in Marine Diatoms. PLoS ONE 2008, 3, e2896. [Google Scholar] [CrossRef] [PubMed]
- Eilers, U.; Dietzel, L.; Breitenbach, J.; Büchel, C.; Sandmann, G. Identification of Genes Coding for Functional Zeaxanthin Epoxidases in the Diatom Phaeodactylum Tricornutum. J. Plant Physiol 2016, 192, 64–70. [Google Scholar] [CrossRef]
- Dautermann, O.; Lohr, M. A Functional Zeaxanthin Epoxidase from Red Algae Shedding Light on the Evolution of Light-harvesting Carotenoids and the Xanthophyll Cycle in Photosynthetic Eukaryotes. Plant J 2017, 92, 879–891. [Google Scholar] [CrossRef] [PubMed]
- Karas, B.J.; Diner, R.E.; Lefebvre, S.C.; McQuaid, J.; Phillips, A.P.R.; Noddings, C.M.; Brunson, J.K.; Valas, R.E.; Deerinck, T.J.; Jablanovic, J.; et al. Designer Diatom Episomes Delivered by Bacterial Conjugation. Nat Commun 2015, 6. [Google Scholar] [CrossRef]
- Dima, O.; Inzé, D. The Role of Scientists in Policy Making for More Sustainable Agriculture. Current Biology 2021, 31, R218–R220. [Google Scholar] [CrossRef] [PubMed]
- Voigt, B. EU Regulation of Gene-Edited Plants—A Reform Proposal. Front. Genome Ed. 2023, 5, 1119442. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Zhang, Q.; Jiang, M.; Huang, J.; Yu, L.; Traw, M.B.; Tian, D.; Hurst, L.D.; Yang, S. Mitotic Gene Conversion Can Be as Important as Meiotic Conversion in Driving Genetic Variability in Plants and Other Species without Early Germline Segregation. PLoS Biol 2021, 19, e3001164. [Google Scholar] [CrossRef] [PubMed]
- Nymark, M.; Finazzi, G.; Volpe, C.; Serif, M.; Fonseca, D. de M.; Sharma, A.; Sanchez, N.; Sharma, A.K.; Ashcroft, F.; Kissen, R.; et al. Loss of CpFTSY Reduces Photosynthetic Performance and Affects Insertion of PsaC of PSI in Diatoms. Plant Cell Physiol 2023, pcad014. [Google Scholar] [CrossRef]
- Nymark, M.; Volpe, C.; Hafskjold, M.C.G.; Kirst, H.; Serif, M.; Vadstein, O.; Bones, A.M.; Melis, A.; Winge, P. Loss of ALBINO3b Insertase Results in Truncated Light-Harvesting Antenna in Diatoms. Plant Physiol 2019, 181, 1257–1276. [Google Scholar] [CrossRef]
- Sharma, A.K.; Nymark, M.; Flo, S.; Sparstad, T.; Bones, A.M.; Winge, P. Simultaneous Knockout of Multiple LHCF Genes Using Single SgRNAs and Engineering of a High-Fidelity Cas9 for Precise Genome Editing in Marine Algae. Plant Biotechnol J 2021, 19, 1658–1669. [Google Scholar] [CrossRef] [PubMed]
- Niyogi, K.K.; Bjorkman, O.; Grossman, A.R. Chlamydomonas Xanthophyll Cycle Mutants Identified by Video Imaging of Chlorophyll Fluorescence Quenching. Plant Cell 1997, 1369–1380. [Google Scholar] [CrossRef]
- Jin, E.; Feth, B.; Melis, A. A Mutant of the Green Alga Dunaliella Salina Constitutively Accumulates Zeaxanthin under All Growth Conditions. Biotech & Bioengineering 2003, 81, 115–124. [Google Scholar] [CrossRef]
- Grouneva, I.; Jakob, T.; Wilhelm, C.; Goss, R. A New Multicomponent NPQ Mechanism in the Diatom Cyclotella Meneghiniana. Plant Cell Physiol. 2008, 49, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Lavaud, J.; Materna, A.C.; Sturm, S.; Vugrinec, S.; Kroth, P.G. Silencing of the Violaxanthin De-Epoxidase Gene in the Diatom Phaeodactylum Tricornutum Reduces Diatoxanthin Synthesis and Non-Photochemical Quenching. PLoS ONE 2012, 7, e36806. [Google Scholar] [CrossRef] [PubMed]
- Goss, R.; Ann Pinto, E.; Wilhelm, C.; Richter, M. The Importance of a Highly Active and DeltapH-Regulated Diatoxanthin Epoxidase for the Regulation of the PS II Antenna Function in Diadinoxanthin Cycle Containing Algae. J Plant Physiol 2006, 163, 1008–1021. [Google Scholar] [CrossRef] [PubMed]
- Lavaud, J.; Kroth, P.G. In Diatoms, the Transthylakoid Proton Gradient Regulates the Photoprotective Non-Photochemical Fluorescence Quenching beyond Its Control on the Xanthophyll Cycle. Plant Cell Physiol 2006, 47, 1010–1016. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Applying and Improving AlphaFold at CASP14. Proteins 2021, 89, 1711–1721. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Bertoni, D.; Magana, P.; Paramval, U.; Pidruchna, I.; Radhakrishnan, M.; Tsenkov, M.; Nair, S.; Mirdita, M.; Yeo, J.; et al. AlphaFold Protein Structure Database in 2024: Providing Structure Coverage for over 214 Million Protein Sequences. Nucleic Acids Res 2024, 52, D368–D375. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-Pdb Viewer: An Environment for Comparative Protein Modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol Biol Evol 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Nymark, M.; Sharma, A.K.; Sparstad, T.; Bones, A.M.; Winge, P. A CRISPR/Cas9 System Adapted for Gene Editing in Marine Algae. Sci Rep-Uk 2016, 6. [Google Scholar] [CrossRef]
- Nymark, M.; Sharma, A.K.; Hafskjold, M.C.; Sparstad, T.; Bones, A.M.; Winge, P. CRISPR/Cas9 Gene Editing in the Marine Diatom Phaeodactylum Tricornutum. Bio-protocol 2017, 7, e2442. [Google Scholar] [CrossRef] [PubMed]
- Guillard, R.R.L. Culture of Phytoplankton for Feeding Marine Invertebrates. In Culture of marine invertebrate animals: Proceedings — 1st conference on culture of marine invertebrate animals Greenport; Smith, W.L., Chanley, M.H., Eds.; Springer US: Boston, MA, 1975; pp. 29–60 ISBN 978-1-4615-8714-9.
- Volpe, C.; Vadstein, O.; Andersen, G.; Andersen, T. Nanocosm: A Well Plate Photobioreactor for Environmental and Biotechnological Studies. Lab Chip 2021, 21, 2027–2039. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Al-Dahhan, M.H. Analyzing and Modeling of Photobioreactors by Combining First Principles of Physiology and Hydrodynamics. Biotech & Bioengineering 2004, 85, 382–393. [Google Scholar] [CrossRef]
- Rodriguez, F.; Chauton, M.; Johnsen, G.; Andresen, K.; Olsen, L.M.; Zapata, M. Photoacclimation in Phytoplankton: Implications for Biomass Estimates, Pigment Functionality and Chemotaxonomy. Mar Biol 2006, 148, 963–971. [Google Scholar] [CrossRef]

| LL | HL | PBR | |
|---|---|---|---|
| WT | 1.21 ± 0.17 | 1.80 ± 0.30 | 1.65 ± 0.28 |
| zep2-1 | 1.25 ± 0.14 | 1.71 ± 0.26 | 1.34 ± 0.16 |
| zep2-2 | 1.29 ± 0.12 | 1.86 ± 0.12 | 1.52 ± 0.22 |
| zep3-1 | 1.25 ± 0.06 | 1.76 ± 0.37 | 1.45 ± 0.14 |
| zep3-2 | 1.23 ± 0.05 | 1.78 ± 0.31 | 1.28 ± 0.08 |
| Oligo or primer name | Orientation | Sequence (5’→3’) | Purpose |
|---|---|---|---|
| ZEP2-PAM2_F | Forward | TCGAGCGCGTGGAGATACGGAGAG | Adapter for sgRNA |
| ZEP2-PAM2_R | Reverse | AAACCTCTCCGTATCTCCACGCGC | |
| ZEP3-PAM2_F | Forward | TCGAAGTACGGTAACATCCATGGT | Adapter for sgRNA |
| ZEP3-PAM2_R | Reverse | AAACACCATGGATGTTACCGTACT | |
| ZEP2-PAM12_scrF | Forward | GAATCGATCTGAATTGGCTACG | screening for zep2 (508 bp amplicon) |
| ZEP2-PAM12_scrR | Reverse | CGGTGAAAGTGAACTTGTCCAT | |
| ZEP3-PAM2_scrF | Forward | GCACCACCTTCGAGCAATGT | Screening for zep3 (643 bp amplicon) |
| ZEP3-PAM2_scrR | Reverse | TCGCCAGCGAAAACCGTGTA | |
| ZEP2-PAM2_hrmF | Forward | CTCCGGAAGACGTTGCCTTTGA | HRM for zep2 (146 bp amplicon) |
| ZEP2-PAM2_hrmR | Reverse | TCTCGTACACCGTCACGTCGAA | |
| ZEP3-PAM2_hrmF | Forward | TGGTCTTTCCTTGGCCAAGGTT | HRM for zep3 (111 bp amplicon) |
| ZEP3-PAM2_hrmR | Reverse | GTTACTAGCGAGCTGGATGGGT | |
| M13-rev (-29) | Reverse | CAGGAAACAGCTATGAC | Sequencing primer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





