Submitted:
11 March 2024
Posted:
11 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
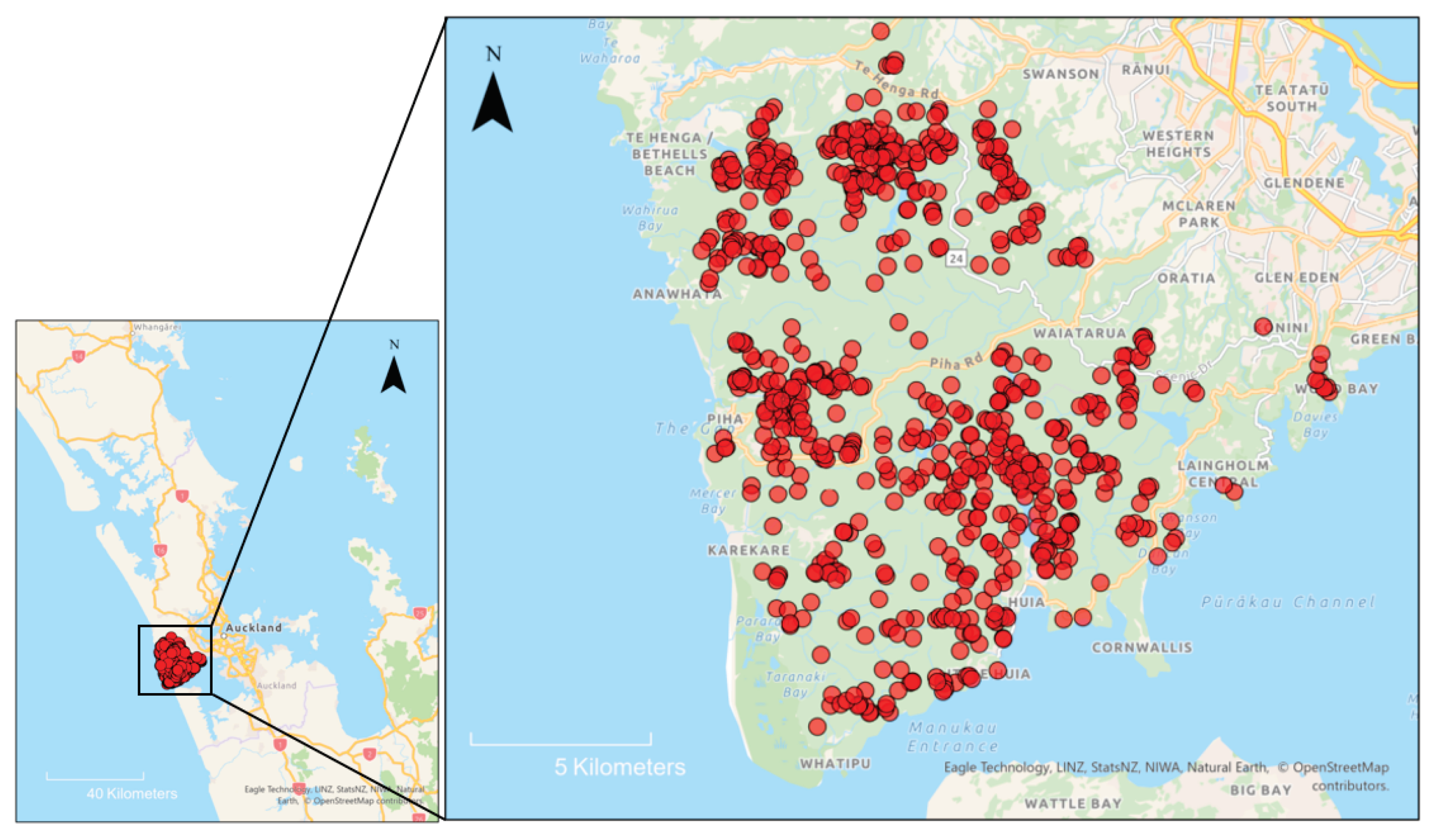
2.1. Site Characteristics and Kauri Dieback History
2.2. Survey Design and Sampling
2.3. Soil Baiting
2.4. Sequencing of Cultures
2.5. Soil DNA Extraction and PCR Amplification
2.6. Controls for Illumina Sequencing of ITS1 Gene Region
2.8. Bioinformatics
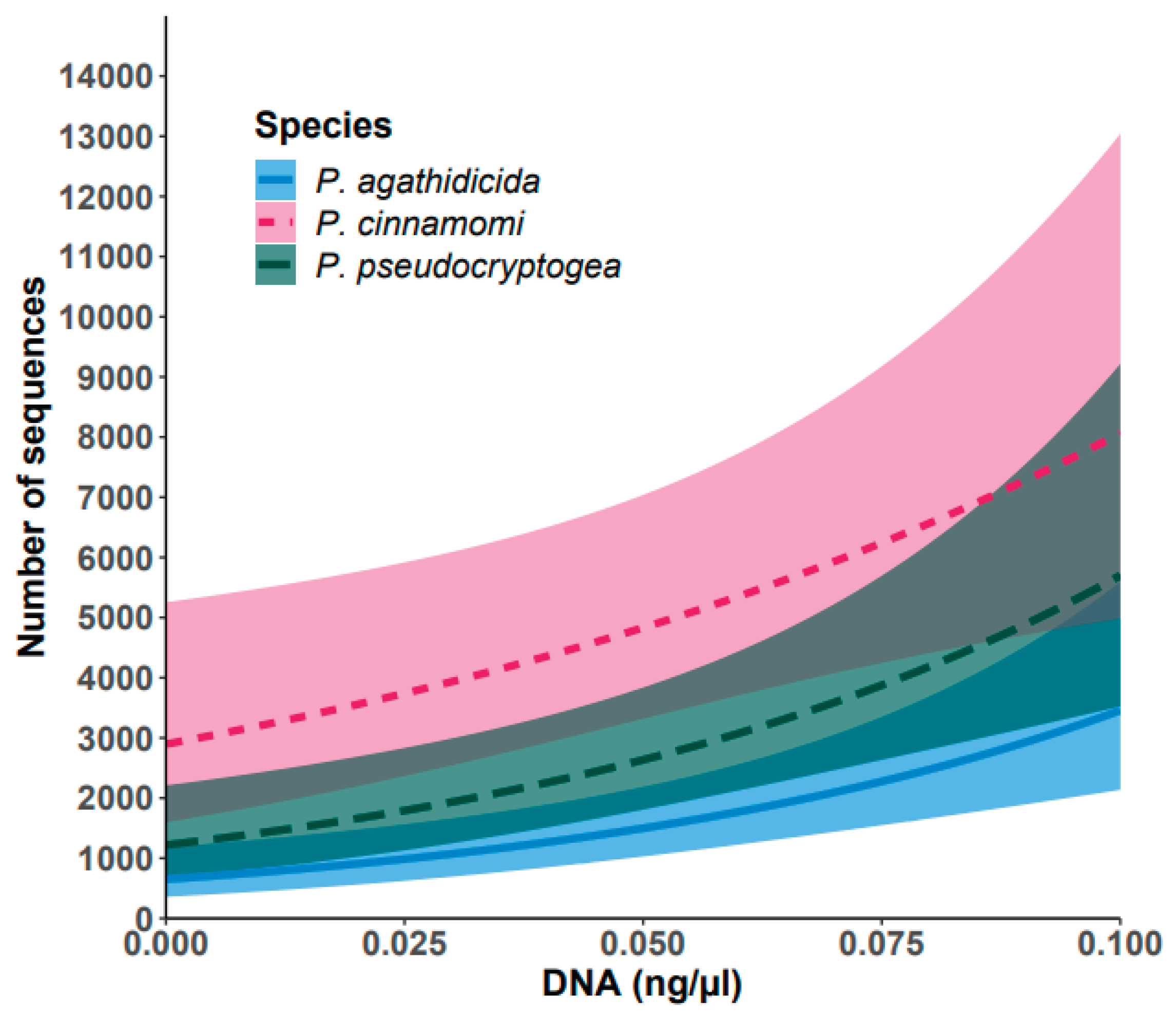
2.9. Data Analyses
3. Results
3.1. Characteristics of Sampled Trees
3.2. Identification of Phytophthora Species by Soil Baiting
3.3. Sequencing Output and Performance of Control Reactions
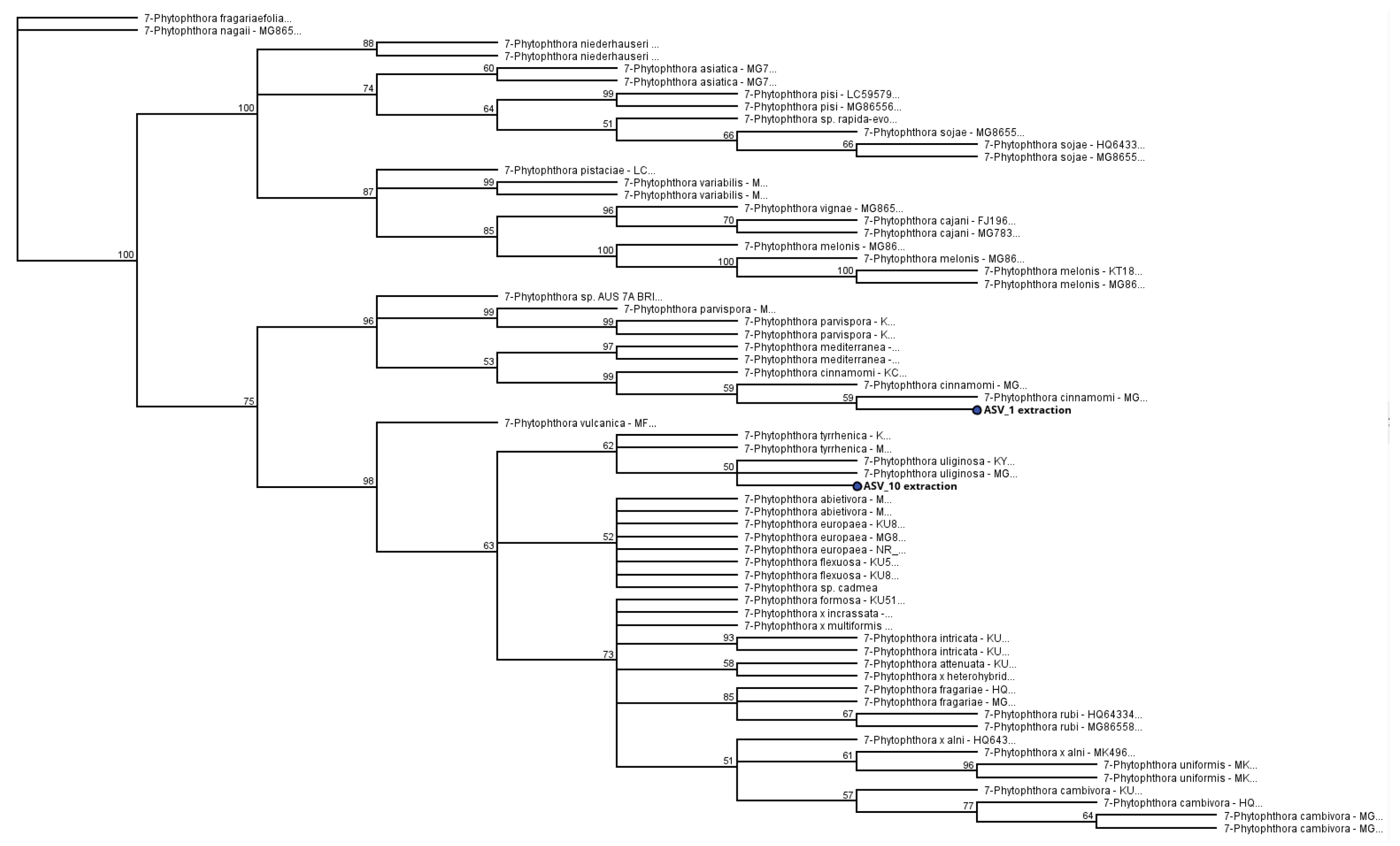
3.4. Identification of Phytophthora Phylotypes by NGS
3.4.1. Unknown Phytophthora Species in Clade 7
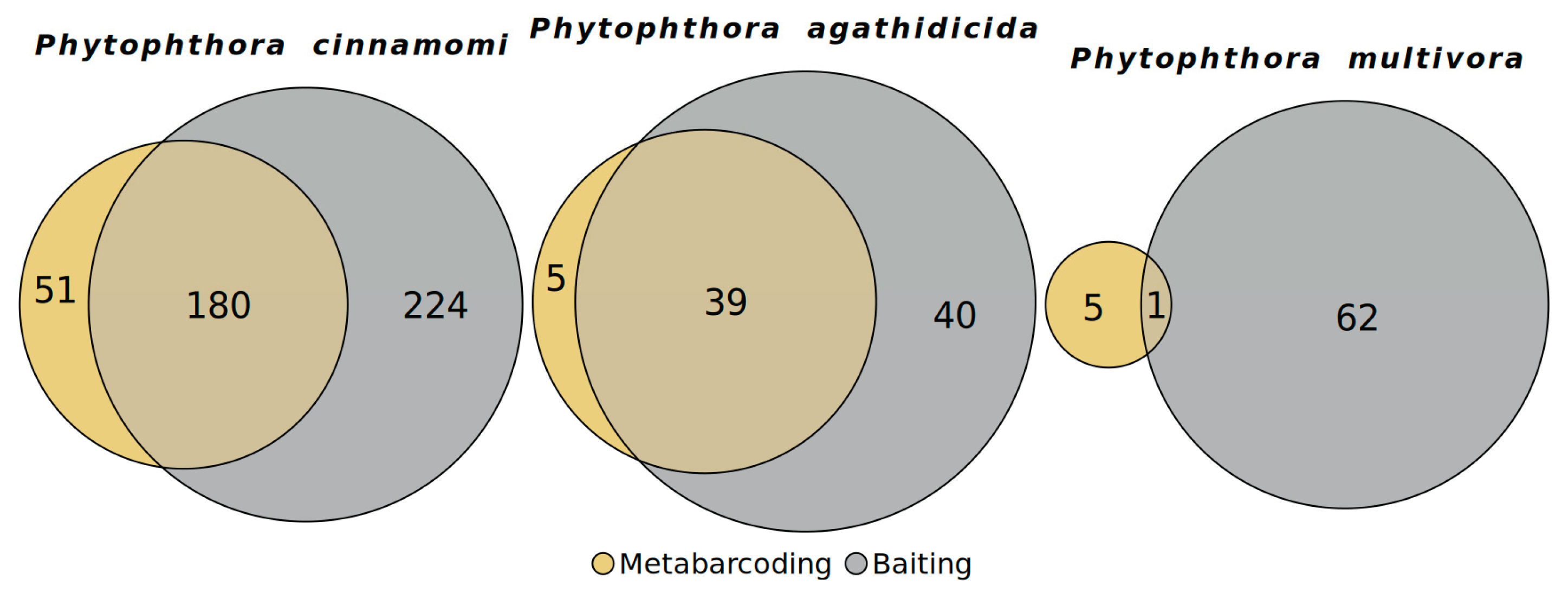
3.5. Comparison of Baiting to Metabarcoding
3.6. Phytophthora Community
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sample | ASV | Reads | Database | Similarity | Query cover | Most similar species |
|---|---|---|---|---|---|---|
| 5359 | 24 | 24937 | ITS1 ref | 100% | 100% | Phytophthora_AUS_1A_KY110340 |
| wgs (whole seq) | 93.36% | 100% | Phytophthora taxon totara | |||
| 5480 | 26 | 19767 | ITS1 ref | 99.50% | 100% | Phytophthora sp. in clade 12A |
| wgs (whole seq) | 99.05% | 100% | P. tubulina | |||
| 5129 | 81 | 3105 | ITS1 ref | 100% | 100% | P. gregata/gibbosa/gonapodyides |
| wgs (whole seq) | 98.84% | 100% | P. chlamydospora |
References
- Hansen, E.M. Phytophthora Species Emerging as Pathogens of Forest Trees. Curr Forestry Rep 2015, 1, 16–24. [Google Scholar] [CrossRef]
- Hansen, E.M.; Reeser, P.W.; Sutton, W. Phytophthora beyond agriculture. Annu Rev Phytopathol 2012, 50, 359–378. [Google Scholar] [CrossRef] [PubMed]
- Prospero, S.; Heinz, M.; Augustiny, E.; Chen, Y.Y.; Engelbrecht, J.; Fonti, M.; Hoste, A.; Ruffner, B.; Sigrist, R.; Van Den Berg, N. Distribution, causal agents, and infection dynamic of emerging ink disease of sweet chestnut in Southern Switzerland. Environ Microbiol 2023. [Google Scholar] [CrossRef] [PubMed]
- Burgess, T.I.; McDougall, K.L.; Scott, P.M.; Hardy, G.E.S.; Garnas, J. Predictors of Phytophthora diversity and community composition in natural areas across diverse Australian ecoregions. Ecography 2019, 42, 594–607. [Google Scholar] [CrossRef]
- Aurangzeb, W.; Guidoni, L.; Rodríguez, C.M.; Cecca, D.; Vannini, A. Exploring the diversity of Phytophthora spp. and the role of Phytophthora multivora in cork and holm oak coastal forests in Italy. Mycol Prog 2023, 22, 51. [Google Scholar] [CrossRef]
- Riddell, C.; Frederickson-Matika, D.; Armstrong, A.; Elliot, M.; Forster, J.; Hedley, P.; Morris, J.; Thorpe, P.; Cooke, D.; Pritchard, L. Metabarcoding reveals a high diversity of woody host-associated Phytophthora spp. in soils at public gardens and amenity woodlands in Britain. PeerJ 2019. [Google Scholar] [CrossRef] [PubMed]
- Legeay, J.; Husson, C.; Boudier, B.; Louisanna, E.; Baraloto, C.; Schimann, H.; Marcais, B.; Buee, M. Surprising low diversity of the plant pathogen Phytophthora in Amazonian forests. Environ Microbiol 2020, 22, 5019–5032. [Google Scholar] [CrossRef] [PubMed]
- Riolo, M.; Aloi, F.; La Spada, F.; Sciandrello, S.; Moricca, S.; Santilli, E.; Pane, A.; Cacciola, S.O. Diversity of Phytophthora Communities across Different Types of Mediterranean Vegetation in a Nature Reserve Area. Forests 2020, 11, 853. [Google Scholar] [CrossRef]
- La Spada, F.; Cock, P.J.A.; Randall, E.; Pane, A.; Cooke, D.E.L.; Cacciola, S.O. DNA Metabarcoding and Isolation by Baiting Complement Each Other in Revealing Phytophthora Diversity in Anthropized and Natural Ecosystems. Journal of Fungi 2022, 8, 330. [Google Scholar] [CrossRef]
- Riit, T.; Cleary, M.; Adamson, K.; Blomquist, M.; Burokienė, D.; Marčiulynienė, D.; Oliva, J.; Poimala, A.; Redondo, M.A.; Strømeng, G.M. , et al. Oomycete Soil Diversity Associated with Betula and Alnus in Forests and Urban Settings in the Nordic–Baltic Region. Journal of Fungi 2023, 9, 926. [Google Scholar] [CrossRef]
- Jung, T.; Blaschke, H.; Oswald, W. Involvement of soilbourne Phytophthora species in central European oak decline and the effect of site factors on the disease. Plant Pathology 2000, 49, 706–718. [Google Scholar] [CrossRef]
- Jung, T. Beech decline in Central Europe driven by the interaction between Phytophthora infections and climatic extremes. Forest Pathology 2009, 39, 73–94. [Google Scholar] [CrossRef]
- Podger, F.D. Phytophthora cinnamomi, a cause of lethal disease in indigenous plant communities in Western Australia. Phytopathology 1972, 62, 972–981. [Google Scholar] [CrossRef]
- Rea, A.J.; Jung, T.; Burgess, T.I.; Stukely, M.J.C.; Hardy, G.E.S.J. Phytophthora elongata sp. nov., a novel pathogen from the Eucalyptus marginata forest of Western Australia. Australas Plant Path 2010, 39, 477–491. [Google Scholar] [CrossRef]
- Ellison, A.M.; Bank, M.S.; Clinton, B.D.; Colburn, E.A.; Elliott, K.; Ford, C.R.; Foster, D.R.; Kloeppel, B.D.; Knoepp, J.D.; Lovett, G.M. , et al. Loss of Foundation Species: Consequences for the Structure and Dynamics of Forested Ecosystems. Front Ecol Environ 2005, 3, 479–486. [Google Scholar] [CrossRef]
- Weir, B.S.; Paderes, E.P.; Anand, N.; Uchida, J.Y.; Pennycook, S.R.; Bellgard, S.E.; Beever, R.E. A taxonomic revision of Phytophthora Clade 5 including two new species, Phytophthora agathidicida and P. cocois. Phytotaxa 2015, 205, 21. [Google Scholar] [CrossRef]
- Beever, R.E.; Waipara, N.; Ramsfield, T.; Dick, M.; Horner, I. Kauri (Agathis australis) under threat from Phytophthora? In Proceedings of the 4th IUFRO Working Party on Phytophthoras in Forests and Native Ecosystems, Monetrey, California, U.S.A, pp 72 - 85. Monetrey, California, U.S.A, 2009. [Google Scholar]
- Waipara, N.W.; Hill, S.; Hill, L.M.W.; Hough, E.G.; Horner, I.J. Surveillance methods to determine tree health, distribution of kauri dieback disease and associated pathogens. New Zealand Plant Prot. 2013, 66, 235–241. [Google Scholar] [CrossRef]
- Winkworth, R.C.; Bellgard, S.E.; McLenachan, P.A.; Lockhart, P.J. The mitogenome of Phytophthora agathidicida: Evidence for a not so recent arrival of the “kauri killing” Phytophthora in New Zealand. PLOS ONE 2021, 16, e0250422. [Google Scholar] [CrossRef]
- Ahmed, M.; Ogden, J. Population dynamics of the emergent conifer Agathis australis (D. Don) Lindl. (Kauri) in New Zealand I. Population structures and tree growth rates in mature stands. New Zealand Journal of Botany 1987, 25, 217–229. [Google Scholar] [CrossRef]
- Steward, G.A.; Beveridge, A.E. A review of New Zealand kauri (Agathis Australis (D.Don) Lindl.): Its ecology, history, growth and potential for management for timber. New Zealand Journal of Forestry Science 2010, 40, 33–59. [Google Scholar]
- Halkett, J.C. A basis for the management of New Zealand kauri (Agathis australis (D. Don) Lindl.) forest. New Zealand Journal of Forestry 1983, 28, 15–23. [Google Scholar]
- Black, A.; Waipara, N.W.; Gerth, M. Correspondence: Save Maori people’s sacred tree species. Nature 2018, 561, 177. [Google Scholar] [CrossRef] [PubMed]
- Fadiman, M. Kauri (Agathis Australis) ethnobotany: Identity, conservation and connection in New Zealand. Florida Geographer 2010, 4–21. [Google Scholar]
- Wyse, S.V.; Burns, B.R.; Wright, S.D. Distinctive vegetation communities are associated with the long-lived conifer Agathis australis (New Zealand kauri, Araucariaceae) in New Zealand rainforests. Austral Ecol 2014, 39, 388–400. [Google Scholar] [CrossRef]
- Padamsee, M.; Johansen, R.B.; Stuckey, S.A.; Williams, S.E.; Hooker, J.E.; Burns, B.R.; Bellgard, S.E. The arbuscular mycorrhizal fungi colonising roots and root nodules of New Zealand kauri Agathis australis. Fungal Biol 2016, 120, 807–817. [Google Scholar] [CrossRef]
- Ogden, J. The long-term conservation of forest diversity in New Zealand. Pacific Conservation Biology 1995, 2, 77–90. [Google Scholar] [CrossRef]
- Beever, R.E.; Bellgard, S.E. ; A., D.M.; Horner, I.J., Ramsfield, T.D. Detection of Phytophthora taxon Agathis, Eds.; Ministry for Agriculture & Forestry, Biosecurity New Zealand: New Zealand, 2010. [Google Scholar]
- Randall, S.D. Fishing for Phytophthora: A yearlong investigation into the diversity of Phytophthora species in the Waitakere Ranges, Auckland, NZ. MSc, University of Auckland, Auckland, New Zealand, 2011.
- Podger, F.D.; Newhook, F.J. Phytophthora cinnamomi in indigenous plant communities in New Zealand. New Zealand Journal of Botany 1971, 9, 625–638. [Google Scholar] [CrossRef]
- Horner, I.J. The role of Phytophthora cinnamomi and other fungal pathogens in the establishment of kauri and kahikatea. University of Auckland, Auckland, New Zealand, 1984.
- NZOR. New Zealand Organisms Register. Available online: https://www.nzor.org.nz/.
- Horner, I.J.; Hough, E.G. Pathogenicity of four Phytophthora species on kauri: in vitro and glasshouse trials. New Zealand Plant Prot. 2014, 67, 54–59. [Google Scholar] [CrossRef]
- Català, S.; Pérez-Sierra, A.; Abad-Campos, P. The Use of Genus-Specific Amplicon Pyrosequencing to Assess Phytophthora Species Diversity Using eDNA from Soil and Water in Northern Spain. PLOS ONE 2015, 10, e0119311. [Google Scholar] [CrossRef]
- Burgess, T.I.; White, D.; McDougall, K.M.; Garnas, J.; Dunstan, W.A.; Català, S.; Carnegie, A.J.; Worboys, S.; Cahill, D.; Vettraino, A.M. , et al. Distribution and diversity of Phytophthora across Australia. Pacific Conservation Biology 2017, 23, 150–162. [Google Scholar] [CrossRef]
- Català, S.; Berbegal, M.; Pérez-Sierra, A.; Abad-Campos, P. Metabarcoding and development of new real-time specific assays reveal Phytophthora species diversity in holm oak forests in eastern Spain. Plant Pathology 2017, 66, 115–123. [Google Scholar] [CrossRef]
- Khaliq, I.; Hardy, G.; White, D.; Burgess, T.I. eDNA from roots: a robust tool for determining Phytophthora communities in natural ecosystems. FEMS Microbiol. Ecol. 2018, 94, 11. [Google Scholar] [CrossRef] [PubMed]
- Bose, T.; Wingfield, M.J.; Roux, J.; Vivas, M.; Burgess, T.I. Community composition and distribution of Phytophthora species across adjacent native and non-native forests of South Africa. Fungal Ecol. 2018, 36, 17–25. [Google Scholar] [CrossRef]
- Vannini, A.; Bruni, N.; Tomassini, A.; Franceschini, S.; Vettraino, A.M. Pyrosequencing of environmental soil samples reveals biodiversity of the Phytophthora resident community in chestnut forests. FEMS Microbiol. Ecol. 2013, 85, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Byers, A.K.; Condron, L.; Donavan, T.; O’Callaghan, M.; Patuawa, T.; Waipara, N.; Black, A. Soil microbial diversity in adjacent forest systems - contrasting native, old growth kauri (Agathis australis) forest with exotic pine (Pinus radiata) plantation forest. FEMS Microbiol. Ecol. 2020, 96, 12. [Google Scholar] [CrossRef]
- Byers, A.K.; Condron, L.; O’Callaghan, M.; Waipara, N.; Black, A. The response of soil microbial communities to the infection of kauri (Agathis australis) seedlings with Phytophthora agathidicida. Forest Pathology 2021, 51. [Google Scholar] [CrossRef]
- Jongkind, A.G.; Buurman, P. The effect of kauri (Agathis australis) on grain size distribution and clay mineralogy of andesitic soils in the Waitakere Ranges, New Zealand. Geoderma 2006, 134, 171–186. [Google Scholar] [CrossRef]
- Hill, L.; Stanley, R.; Hammon, C.; Waipara, N. Kauri Dieback Report 2017: An Investigation into the Distribution of Kauri Dieback, and Implications for its Future Management, within the Waitakere Ranges Regional Park; Auckland Council.: Auckland, New Zealand, 2017.
- Froud, K.; Chew, Y.C.; Kean, J.; Jae, M.; Killick, S.; Ashcby, E.; Taua-Gordon, R.; Jamieson, A.; Tolich, L. 2021 Waitakere Ranges Kauri Population Health Monitoring Survey; Auckland Council technical report: 2022.
- Dick, M.; Bellgard, S. Soil survey method for Phytophthora taxon Agathis. Report prepared for the Ministry of Agriculture and Forestry on behalf of Kauri Dieback Joint Agency, Version 2.1, modified by A.J. Beauchamp, Department of Conservation, Pp. 13. 2012. [Google Scholar]
- O’Brien, P.A.; Williams, N.; Hardy, G.E.S. Detecting Phytophthora. Critical Reviews in Microbiology 2009, 35, 169–181. [Google Scholar] [CrossRef]
- Erwin, D.C.; Ribeiro, O.K. Phytophthora diseases worldwide; APS Press: Minnesota, 1996. [Google Scholar]
- White, T.; Bruns, T.; Lee, S.; Taylor, J.; Innis, M.; Gelfand, D.; Sninsky, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. 1990, 31, 315–322. [Google Scholar]
- Robideau, G.P.; De Cock, A.W.A.M.; Coffey, M.D.; Voglmayr, H.; Brouwer, H.; Bala, K.; Chitty, D.W.; Désaulniers, N.; Eggertson, Q.A.; Gachon, C.M.M. , et al. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Molecular Ecology Resources 2011, 11, 1002–1011. [Google Scholar] [CrossRef]
- Legeay, J.; Husson, C.; Cordier, T.; Vacher, C.; Marcais, B.; Buée, M. Comparison and validation of Oomycetes metabarcoding primers for Phytophthora high throughput sequencing. J Plant Pathol 2019, 101, 743–748. [Google Scholar] [CrossRef]
- Scibetta, S.; Schena, L.; Chimento, A.; Cacciola, S.O.; Cooke, D.E.L. A molecular method to assess Phytophthora diversity in environmental samples. J Microbiol Meth 2012, 88, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.N.; Coffey, M.D. Mitochondrial Haplotype Analysis for Differentiation of Isolates of Phytophthora cinnamomi. Phytopathology® 2012, 102, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Foster, Z.S.L.; Albornoz, F.E.; Fieland, V.J.; Larsen, M.M.; Jones, F.A.; Tyler, B.M.; Nguyen, H.D.T.; Burgess, T.I.; Riddell, C.; Voglmayr, H. , et al. A New Oomycete Metabarcoding Method Using the rps10 Gene. Phytobiomes Journal 2022, 6, 214–226. [Google Scholar] [CrossRef]
- Illumina. llumina 16S metagenomic sequencing library preparation (IlluminaTechnical Note 15044223). Accessed 24 Jan 2024 2013. Available online: https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf.
- McDougal, R.; Bellgard, S.E.; Scott, P.; Ganley, B. Comparison of a real time PCR assay and a soil bioassay technique for detection of Phytophthora taxon Agathis from soil; 2014.
- Than, D.; Hughes, K.; Boonhan, N.; Tomlinson, J.; Woodhall, J.; Bellgard, S. A TaqMan real-time PCR assay for the detection of Phytophthora ‘taxon Agathis’ in soil, pathogen of Kauri in New Zealand. Forest Pathology 2013, 43. [Google Scholar] [CrossRef]
- Kunadiya, M.; White, D.; Dunstan, W.A.; St, G.E.; Hardy, J.; Andjic, V.; Burgess, T.I. Pathways to false positive diagnoses using molecular genetic detection methods; Phytophthora cinnamomi a case study. Fems Microbiol Lett 2017, fnx009. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Version 0.12.0 [Online]. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- R Core Team. R: A language and Environment for Statistical Computing. R Foundation for Statistical Computing.: Vienna, Austria., 2023.
- Ripley, B.; Venables, B.; Bates, D.; Hornik, K.; Gebhardt, A.; Firth, D. MASS: Support Functions and Datasets for Venables and Ripley’s MASS. R Package Version 7.45. 2016. [Google Scholar]
- Zuur, A.F.; Ieno, E.N. A protocol for conducting and presenting results of regression-type analyses. Methods in Ecology and Evolution 2016, 7, 636–645. [Google Scholar] [CrossRef]
- Griffith, D.M.; Veech, J.A.; Marsh, C.J. cooccur: Probabilistic Species Co-Occurrence Analysis in R. Journal of Statistical Software, Code Snippets 2016, 69, 1–17. [Google Scholar] [CrossRef]
- Larsson, J. eulerr: Area-Proportional Euler and Venn Diagrams with Ellipses. R package version 7.0.0.. 2022. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 3.3.5; Springer-Verlag: New York, 2016. [Google Scholar]
- Gardner, J.F.; Dick, M.A.; Bader, M.K.-F. Susceptibility of New Zealand flora to Phytophthora kernoviae and its seasonal variability in the field. New Zealand Journal of Forestry Science 2015, 45, 23. [Google Scholar] [CrossRef]
- Scott, P.; Williams, N. Phytophthora diseases in New Zealand forests. New Zealand Journal of Forestry 2014, 59, 14–21. [Google Scholar]
- Bregant, C.; Batista, E.; Hilário, S.; Linaldeddu, B.T.; Alves, A. Phytophthora Species Involved in Alnus glutinosa Decline in Portugal. Pathogens 2023, 12, 276. [Google Scholar] [CrossRef] [PubMed]
- Seddaiu, S.; Brandano, A.; Ruiu, P.A.; Sechi, C.; Scanu, B. An overview of Phytophthora species inhabiting declining Quercus suber stands in Sardinia (Italy). Forests 2020, 11. [Google Scholar] [CrossRef]
- Corcobado, T.; Cech, T.L.; Daxer, A.; Ďatková, H.; Janoušek, J.; Patra, S.; Jahn, D.; Hüttler, C.; Milenković, I.; Tomšovský, M. , et al. Phytophthora, Nothophytophthora and Halophytophthora diversity in rivers, streams and riparian alder ecosystems of Central Europe. Mycol Prog 2023, 22. [Google Scholar] [CrossRef] [PubMed]
- Greslebin, A.G.; Hansen, E.M.; Winton, L.M.; Rajchenberg, M. Phytophthora species from declining Austrocedrus chilensis forests in Patagonia, Argentina. Mycologia 2005, 97, 218–228. [Google Scholar] [CrossRef]
- Lyubenova, A.; Kostov, K.; Tsvetkov, I.; Slavov, S. Possible contribution of Phytophthora spp. in decline of Norway spruce (P. abies (L.) H. Karst.) stands in Vitosha Mountain. Silva Balcanica 2014, 15, 100–109. [Google Scholar]
- Riddell, C.E.; Dun, H.F.; Elliot, M.; Armstrong, A.C.; Clark, M.; Forster, J.; Hedley, P.E.; Green, S. Detection and spread of Phytophthora austrocedri within infected Juniperus communis woodland and diversity of co-associated Phytophthoras as revealed by metabarcoding. Forest Pathology 2020, 50. [Google Scholar] [CrossRef]
- Newhook, F.J. The association of Phytophthora spp. with mortality of Pinus radiata and other conifers. New Zealand Journal of Agricultural Research 1959, 2, 808–843. [Google Scholar] [CrossRef]
- Brien, R.M.; Dingley, J.M. Fourth supplement to “A revised list of plant diseases recorded in New Zealand”, 1957–1958. New Zealand Journal of Agricultural Research 1959, 2, 406–413. [Google Scholar] [CrossRef]
- Scott, P.M.; Burgess, T.I.; Barber, P.A.; Shearer, B.L.; Stukely, M.J.C.; Hardy, G.E.S.; Jung, T. Phytophthora multivora sp nov., a new species recovered from declining Eucalyptus, Banksia, Agonis and other plant species in Western Australia. Persoonia 2009, 22, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tsykun, T.; Prospero, S.; Schoebel, C.N.; Rea, A.; Burgess, T.I. Global invasion history of the emerging plant pathogen Phytophthora multivora. Bmc Genomics 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Burgess, T.I.; Scott, J.K.; McDougall, K.L.; Stukely, M.J.C.; Crane, C.; Dunstan, W.A.; Brigg, F.; Andjic, V.; White, D.; Rudman, T. , et al. Current and projected global distribution of Phytophthora cinnamomi, one of the world’s worst plant pathogens. Global Change Biology 2017, 23, 1661–1674. [Google Scholar] [CrossRef] [PubMed]
- Hardham, A.R.; Blackman, L.M. Phytophthora cinnamomi. Mol Plant Pathol 2018, 19, 260–285. [Google Scholar] [CrossRef]
- Burgess, T.I.; White, D.; Sapsford, S.J. Comparison of Primers for the Detection of Phytophthora (and Other Oomycetes) from Environmental Samples. Journal of Fungi 2022, 8, 980. [Google Scholar] [CrossRef]
- Sarker, S.R.; McComb, J.; Burgess, T.I.; Hardy, G.E.S.J. Timing and abundance of sporangia production and zoospore release influences the recovery of different Phytophthora species by baiting. Fungal Biol-Uk 2021, 125, 477–484. [Google Scholar] [CrossRef]
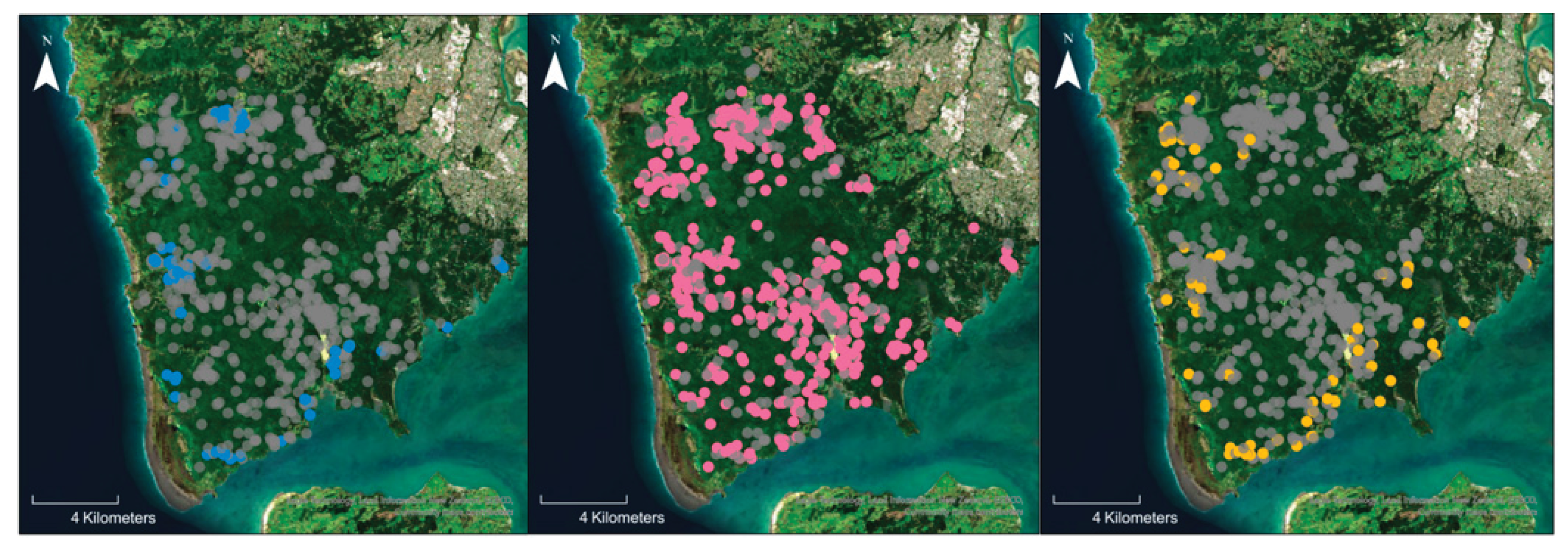
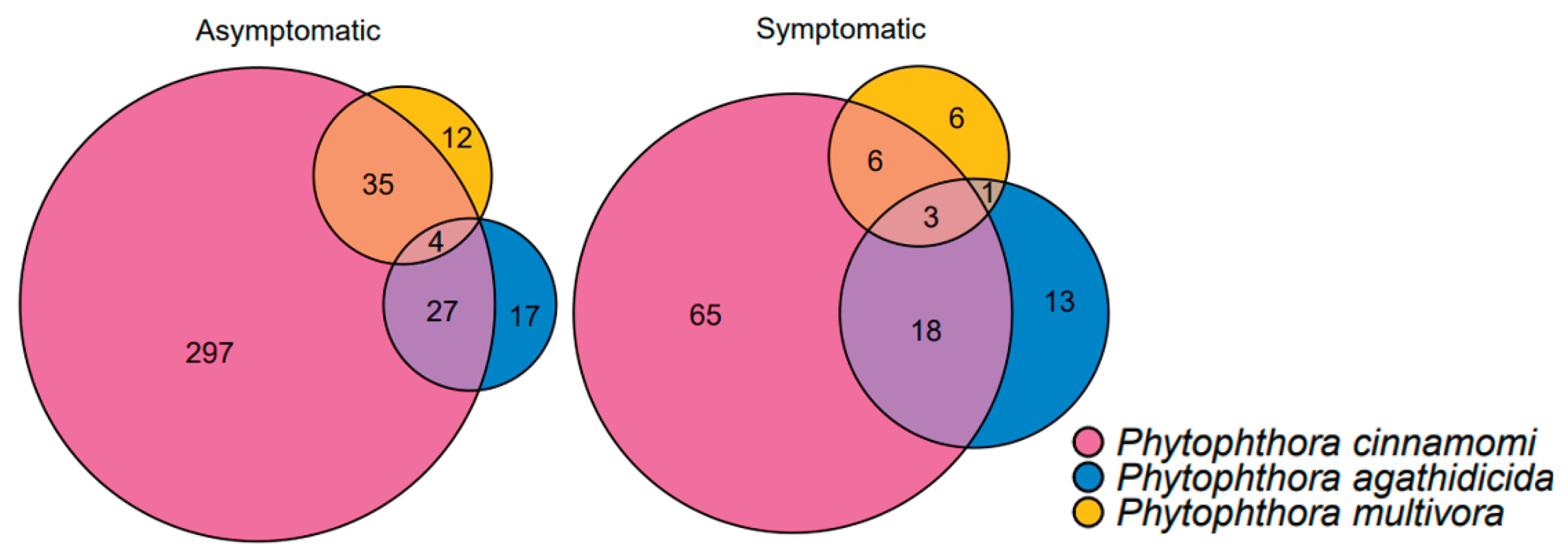
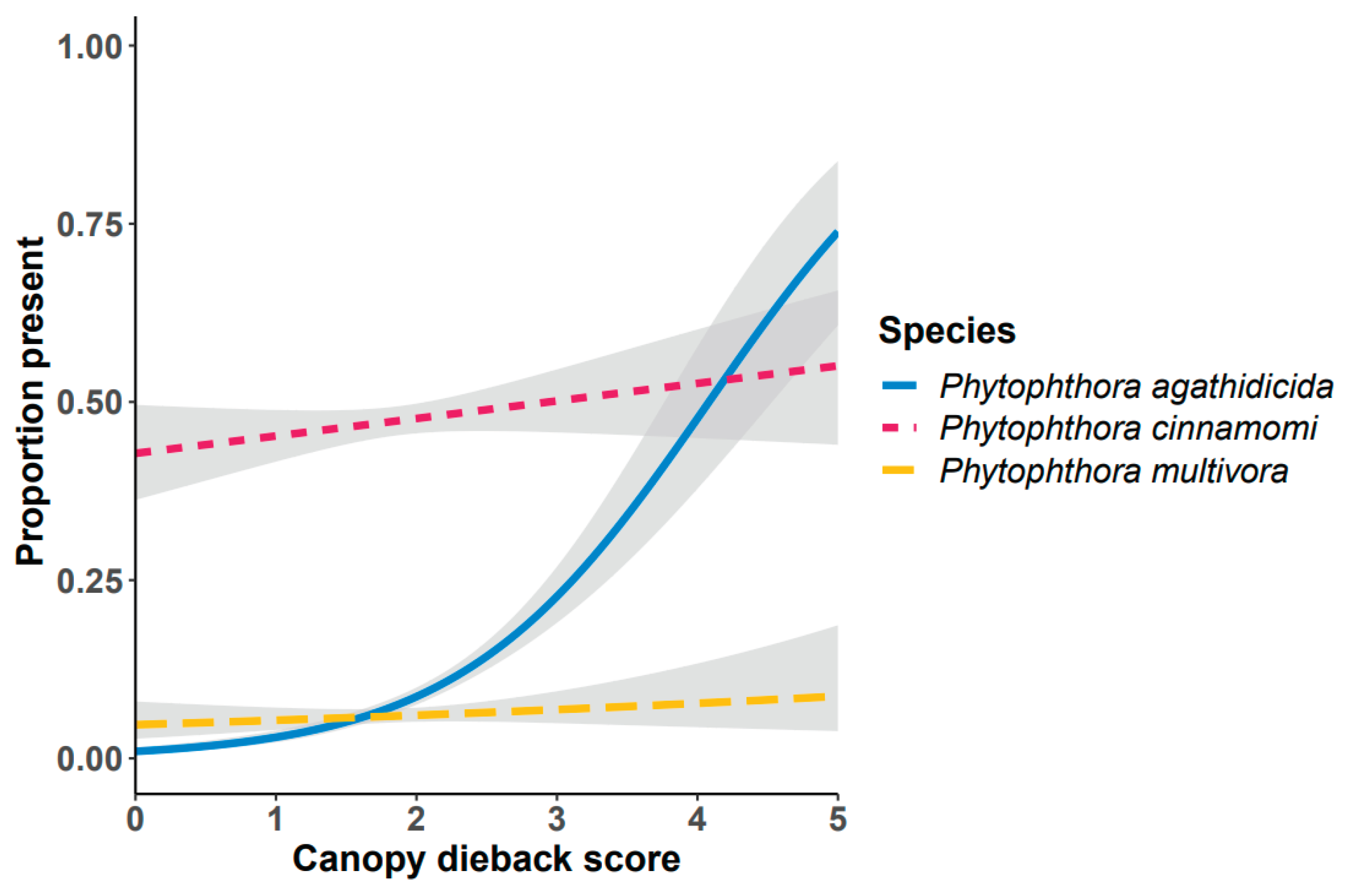
| Primer or probe name | Target species | Sequence (5’ to 3’) | Notes* | Reference |
|---|---|---|---|---|
| ITS_PTA_F2 | P. agathidicida | AACCAATAGTTGGGGGCGA | [55,56] | |
| ITS_PTA_R3 | CTCGCCATGATAGAGCTCGTC | [55] | ||
| ITS_PTA_probe2 | AGCCAAAGCCAGCAGCCG | 5’ FAM, 3’ BHQ1 | [55] | |
| PCIN F6 | P. cinnamomi | CGTGGCGGGCCCTATC | [57] | |
| PCIN R2 | AAAAGAGAGGCTACTAGCTCAGTTCCC | [57] | ||
| PCIN probe | TGGCGAGCGTTTGGGTCCCTCT | 5’ HEX, 3’ BHQ2 | [57] |
| Phytophthora species | Clade | Soil baiting | Metabarcoding (soil eDNA) | Total detections | Total detections (%) |
|---|---|---|---|---|---|
| P. cinnamomi | 7 | 404 | 231 | 455 | 59.3 |
| P. agathidicida | 5 | 79 | 44 | 84 | 11.0 |
| P. multivora | 2 | 63 | 6 | 68 | 8.9 |
| P. sp (P. europaea-like) | 7 | 0 | 20 | 20 | 2.6 |
| P. cactorum/P. aleatoria | 1 | 0 | 4 | 4 | 0.5 |
| P. pseudocryptogea | 8 | 1 | 2 | 2 | 0.3 |
| P. kernoviae | 10b | 0 | 2 | 2 | 0.3 |
| Reads (%)* | |||||
|---|---|---|---|---|---|
| Sample ID | DNA (ng/µl) | P. cinnamomi (H1454) | P. agathidicida (H1453) | P. pseudocryptogea (H1258) | P. multivora (H1467) |
| 6000 | 0.1 | 53.3 | 16.0 | 27.9 | 0.0 |
| 6001 | 0.01 | 51.0 | 19.4 | 27.4 | 0.0 |
| 6002 | 0.001 | 55.9 | 15.3 | 26.8 | 0.7 |
| 6011 | 0.0001 | 60.7 | 2.8 | 33.4 | 0.0 |
| 6003 | 0.01 | - | 0.3 | - | - |
| 0.1 | 58.3 | - | 38.2 | 0.0 | |
| 6004 | 0.001 | - | 31.4 | - | - |
| 0.1 | 16.5 | - | 48.7 | 1.1 | |
| 6005 | 0.01 | 3.3 | - | - | - |
| 0.1 | - | 37.5 | 57.6 | 1.0 | |
| 6006 | 0.001 | 69.6 | - | - | - |
| 0.1 | - | 19.2 | 7.0 | 0.0 | |
| 6007 | 0.01 | - | - | 0.7 | - |
| 0.1 | 71.0 | 22.8 | - | 0.3 | |
| 6008 | 0.001 | - | - | 30.2 | - |
| 0.1 | 52.0 | 17.5 | - | 0.0 | |
| 6009 | 0.01 | - | - | - | 0.0 |
| 0.1 | 53.2 | 17.1 | 29.7 | - | |
| 6010 | 0.001 | - | - | - | 0.0 |
| 0.1 | 62.2 | 9.1 | 26.6 | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





