Submitted:
06 September 2023
Posted:
07 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Materials and DNA Extraction.
2.2. Chloroplast Genome Sequencing and Assembling.
2.3. Repeat Sequences and SSRs.
2.4. Variations and Divergence Hotspot Regions of the cp Genomes.
2.5. Phylogenomic Reconstruction Based on cp Genomes.
3. Results
3.1. Chloroplast Genome Structure
3.2. IR Expansion and Contraction
3.3. Variations and Divergence Hotspot Regions
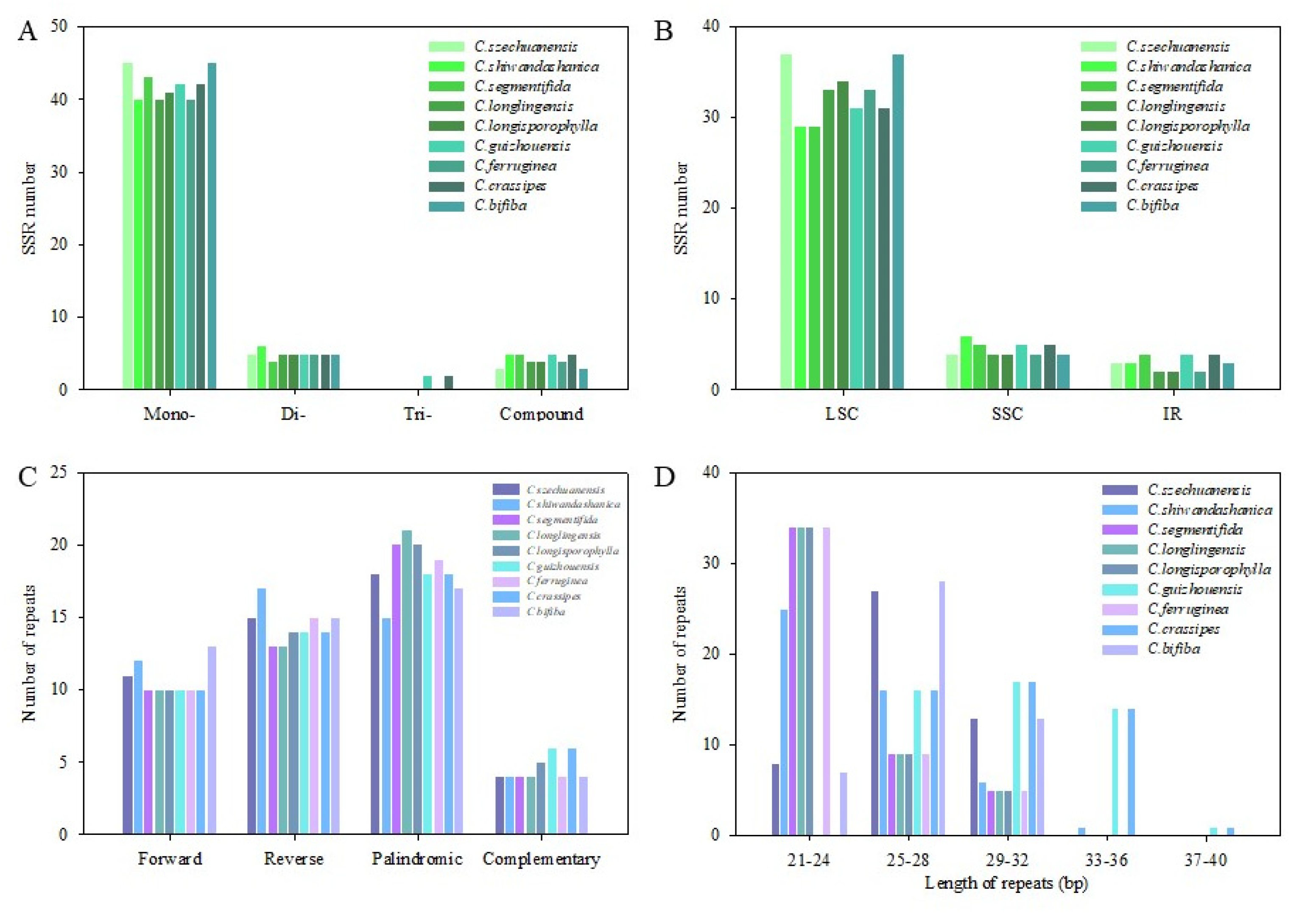
3.4. SSR and Large Repeats
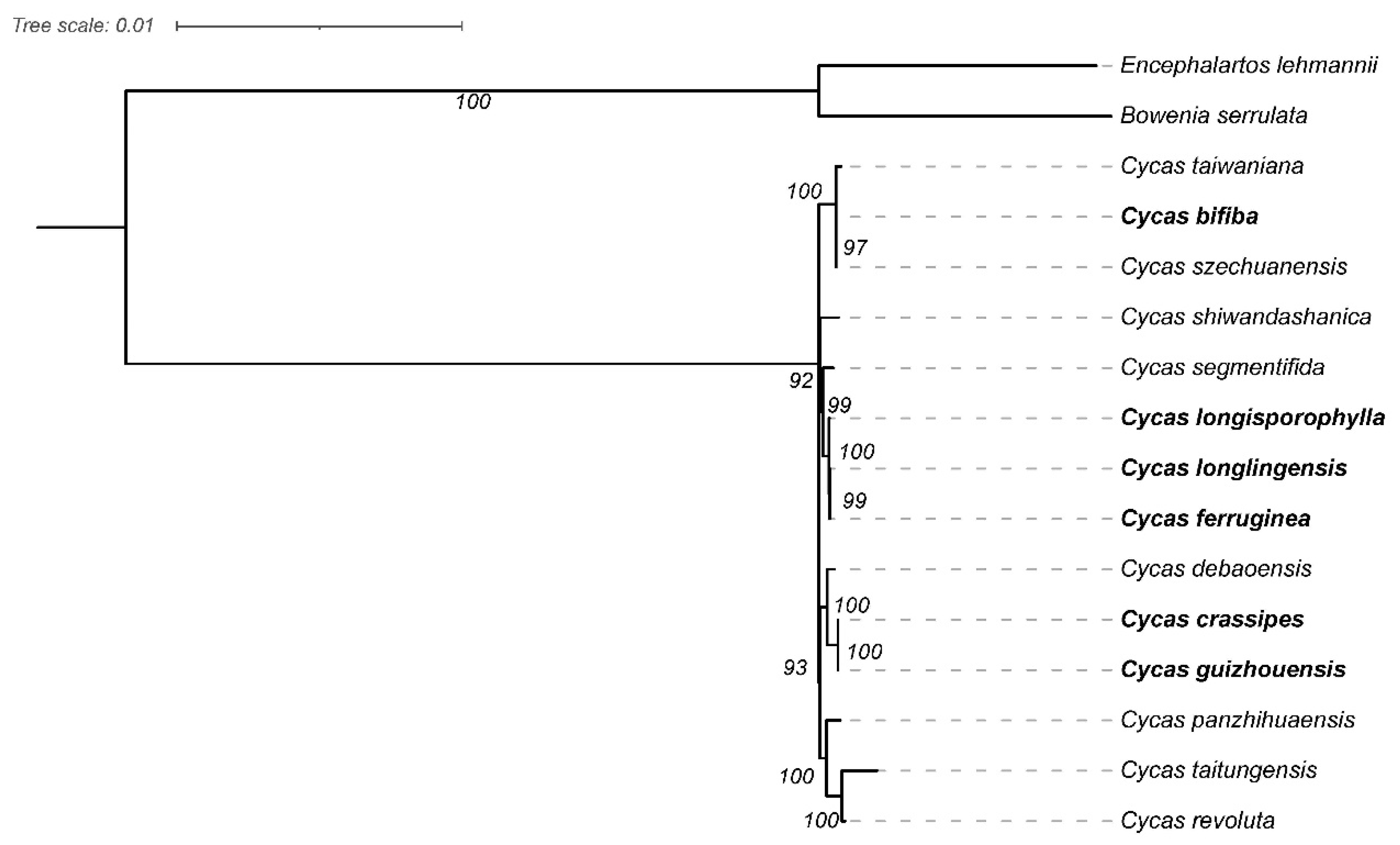
3.5. Phylogenomic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhifeng, G.; Thomas, B.A. A review of fossil cycad megasporophylls, with new evidence of Crossozamia Pomel and its associated leaves from the Lower Permian of Taiyuan, China. Review of Palaeobotany and Palynology 1989, 60, 205-223. [CrossRef]
- Calonje, M.; Stevenson, D.; Stanberg, L. The world list of cycads, online edition. Recuperado el 2019, 10.
- Osborne, R. The world cycad census and a proposed revision of the threatened species status for cycad taxa. Biological Conservation 1995, 71, 1-12. [CrossRef]
- Spiekermann, R.; Jasper, A.; Siegloch, A.M.; Guerra-Sommer, M.; Uhl, D. Not a lycopsid but a cycad-like plant: Iratinia australis gen. nov. et sp. nov. from the Irati Formation, Kungurian of the Paraná Basin, Brazil. Review of Palaeobotany and Palynology 2021, 289, 104415. [CrossRef]
- Liu, J.; Lindstrom, A.J.; Chen, Y.S.; Nathan, R.; Gong, X. Congruence between ocean-dispersal modelling and phylogeography explains recent evolutionary history of Cycas species with buoyant seeds. New Phytologist 2021, 232, 1863-1875. [CrossRef]
- Zheng, Y.; Liu, J.; Feng, X.; Gong, X. The distribution, diversity, and conservation status of Cycas in China. Ecology and Evolution 2017, 7, 3212-3224.
- Tao, Y.; Chen, B.; Kang, M.; Liu, Y.; Wang, J. Genome-wide evidence for complex hybridization and demographic history in a group of Cycas from China. Frontiers in Genetics 2021, 12, 717200. [CrossRef]
- Hill, K. Infrageneric relationships, phylogeny, and biogeography of the genus Cycas (Cycadaeceae). In Proceedings of the Proceedings of the Third International Conference on Cycad Biology, 1995; pp. 139-175.
- Hill, K. Character evolution, species recognition and classification concepts in the Cycadaceae. In Cycad classification: concepts and recommendations; CABI Publishing Wallingford UK: 2004; pp. 23-44.
- Xiao, L.-Q.; Möller, M. Nuclear ribosomal ITS functional paralogs resolve the phylogenetic relationships of a late-Miocene radiation cycad Cycas (Cycadaceae). PloS one 2015, 10, e0117971. [CrossRef]
- Saina, J.K.; Li, Z.-Z.; Gichira, A.W.; Liao, Y.-Y. The complete chloroplast genome sequence of tree of heaven (Ailanthus altissima (Mill.)(Sapindales: Simaroubaceae), an important pantropical tree. International journal of molecular sciences 2018, 19, 929.
- Liu, J.; Zhang, S.; Nagalingum, N.S.; Chiang, Y.-C.; Lindstrom, A.J.; Gong, X. Phylogeny of the gymnosperm genus Cycas L.(Cycadaceae) as inferred from plastid and nuclear loci based on a large-scale sampling: evolutionary relationships and taxonomical implications. Molecular Phylogenetics and Evolution 2018, 127, 87-97. [CrossRef]
- Han, J.; Wang, M.; Qiu, Q.; Guo, R. Characterization of the complete chloroplast genome of Cycas panzhihuaensis. Conservation genetics resources 2017, 9, 21-23. [CrossRef]
- Wang, Y.; Wang, Z.; Zhang, L.; Yang, X. The complete chloroplast genome of Cycas Szechuanensis, an extremely endangered species. Mitochondrial DNA Part B 2018, 3, 974-975. [CrossRef]
- Zhang, D.; Cao, Y.; Lu, Z. The complete chloroplast genome of Cycas bifida, an extremely small population protected species. Mitochondrial DNA Part B 2021, 6, 2960-2961.
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current genetics 2007, 52, 267-274. [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451-1452. [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.i.; Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic acids research 2002, 30, 3059-3066.
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nature methods 2012, 9, 772-772. [CrossRef]
- Posada, D.; Crandall, K.A. MODELTEST: testing the model of DNA substitution. Bioinformatics (Oxford, England) 1998, 14, 817-818. [CrossRef]
- Dong, W.; Xu, C.; Li, W.; Xie, X.; Lu, Y.; Liu, Y.; Jin, X.; Suo, Z. Phylogenetic resolution in Juglans based on complete chloroplast genomes and nuclear DNA sequences. Frontiers in plant science 2017, 8, 1148. [CrossRef]
- Clegg, M.T.; Zurawski, G. Chloroplast DNA and the study of plant phylogeny: present status and future prospects. Molecular systematics of plants 1992, 1-13. [CrossRef]
- Millen, R.S.; Olmstead, R.G.; Adams, K.L.; Palmer, J.D.; Lao, N.T.; Heggie, L.; Kavanagh, T.A.; Hibberd, J.M.; Gray, J.C.; Morden, C.W. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. The Plant Cell 2001, 13, 645-658.
- Ravi, V.; Khurana, J.; Tyagi, A.; Khurana, P. An update on chloroplast genomes. Plant Systematics and Evolution 2008, 271, 101-122.
- Mehmood, F.; Shahzadi, I.; Waseem, S.; Mirza, B.; Ahmed, I.; Waheed, M.T. Chloroplast genome of Hibiscus rosa-sinensis (Malvaceae): comparative analyses and identification of mutational hotspots. Genomics 2020, 112, 581-591. [CrossRef]
- Tang, J.; Zou, R.; Wei, X.; Li, D. Complete Chloroplast Genome Sequences of Five Ormosia Species: Molecular Structure, Comparative Analysis, and Phylogenetic Analysis. Horticulturae 2023, 9, 796.
- Asaf, S.; Khan, A.L.; Khan, A.R.; Waqas, M.; Kang, S.-M.; Khan, M.A.; Lee, S.-M.; Lee, I.-J. Complete chloroplast genome of Nicotiana otophora and its comparison with related species. Frontiers in plant science 2016, 7, 843. [CrossRef]
- Shahzadi, I.; Mehmood, F.; Ali, Z.; Malik, M.S.; Waseem, S.; Mirza, B.; Ahmed, I.; Waheed, M.T. Comparative analyses of chloroplast genomes among three Firmiana species: Identification of mutational hotspots and phylogenetic relationship with other species of Malvaceae. Plant Gene 2019, 19, 100199. [CrossRef]
- Song, W.; Ji, C.; Chen, Z.; Cai, H.; Wu, X.; Shi, C.; Wang, S. Comparative analysis the complete chloroplast genomes of nine Musa species: genomic features, comparative analysis, and phylogenetic implications. Frontiers in Plant Science 2022, 13, 832884. [CrossRef]
- Xie, D.-F.; Yu, Y.; Deng, Y.-Q.; Li, J.; Liu, H.-Y.; Zhou, S.-D.; He, X.-J. Comparative analysis of the chloroplast genomes of the Chinese endemic genus Urophysa and their contribution to chloroplast phylogeny and adaptive evolution. International Journal of Molecular Sciences 2018, 19, 1847. [CrossRef]
- Kelchner, S.A. The evolution of non-coding chloroplast DNA and its application in plant systematics. Annals of the Missouri Botanical Garden 2000, 482-498. [CrossRef]
- Liang, C.; Wang, L.; Lei, J.; Duan, B.; Ma, W.; Xiao, S.; Qi, H.; Wang, Z.; Liu, Y.; Shen, X. A comparative analysis of the chloroplast genomes of four Salvia medicinal plants. Engineering 2019, 5, 907-915. [CrossRef]
- Yaradua, S.S.; Alzahrani, D.A.; Albokhary, E.J.; Abba, A.; Bello, A. Complete chloroplast genome sequence of Justicia flava: genome comparative analysis and phylogenetic relationships among Acanthaceae. BioMed Research International 2019, 2019.
- Yang, Y.; Zhou, T.; Duan, D.; Yang, J.; Feng, L.; Zhao, G. Comparative analysis of the complete chloroplast genomes of five Quercus species. Frontiers in plant science 2016, 7, 959. [CrossRef]
- Cui, Y.; Nie, L.; Sun, W.; Xu, Z.; Wang, Y.; Yu, J.; Song, J.; Yao, H. Comparative and phylogenetic analyses of ginger (Zingiber officinale) in the family Zingiberaceae based on the complete chloroplast genome. Plants 2019, 8, 283. [CrossRef]
- Hill, K. The genus Cycas (Cycadaceae) in China. Telopea 2008, 12, 71-118. [CrossRef]
- Whitelock, L.M. The cycads; Timber Press: 2002.
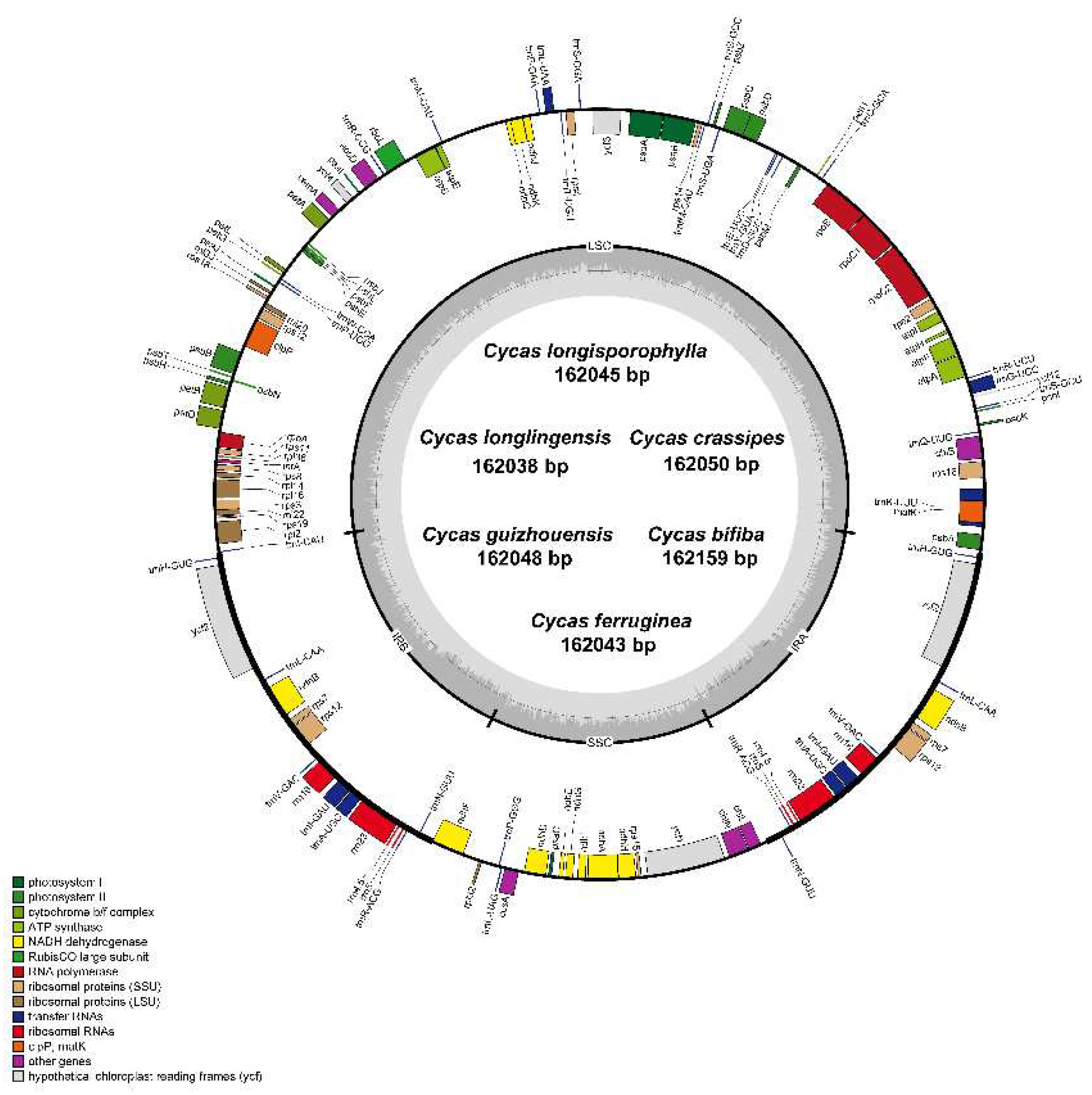
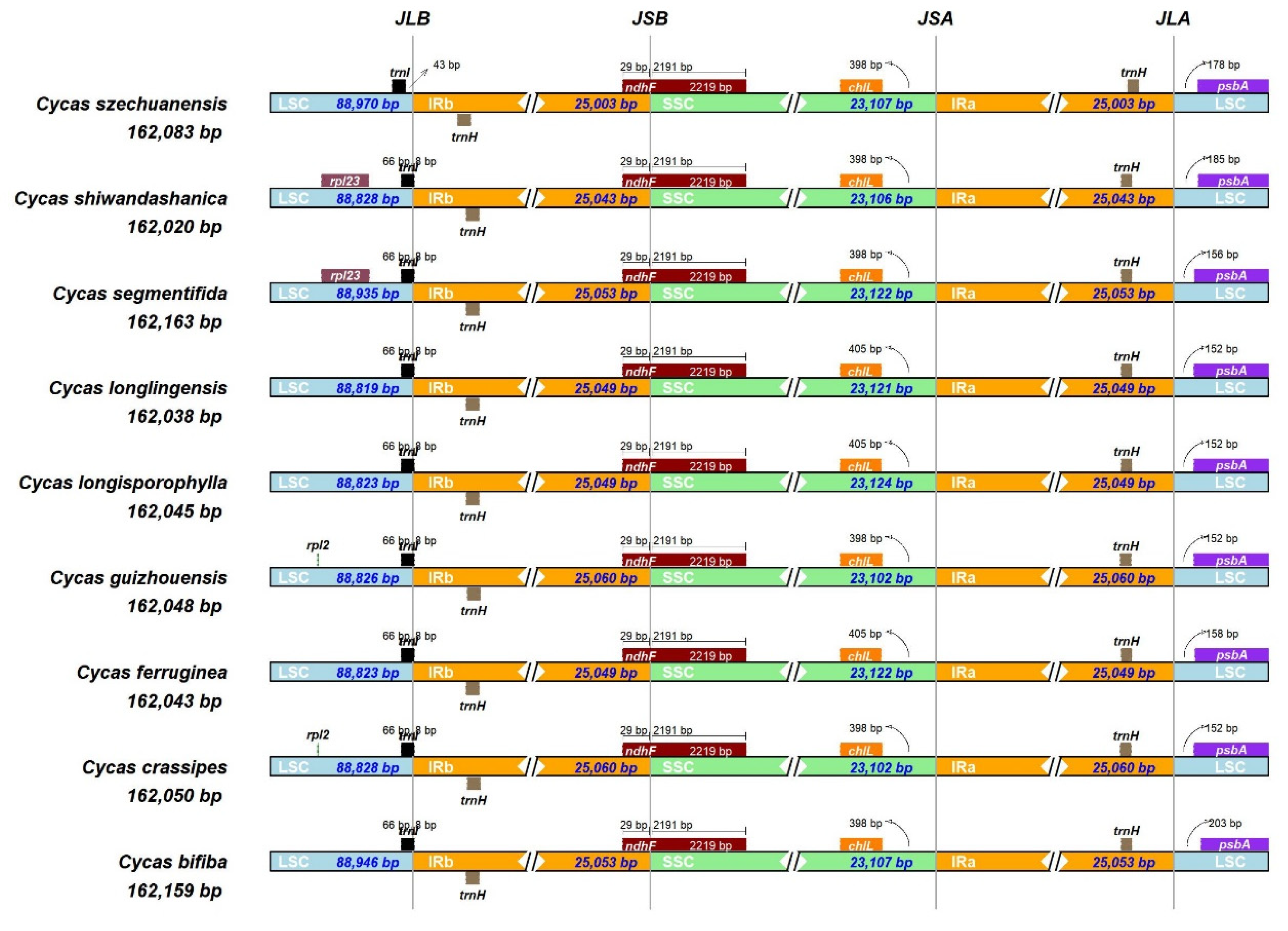
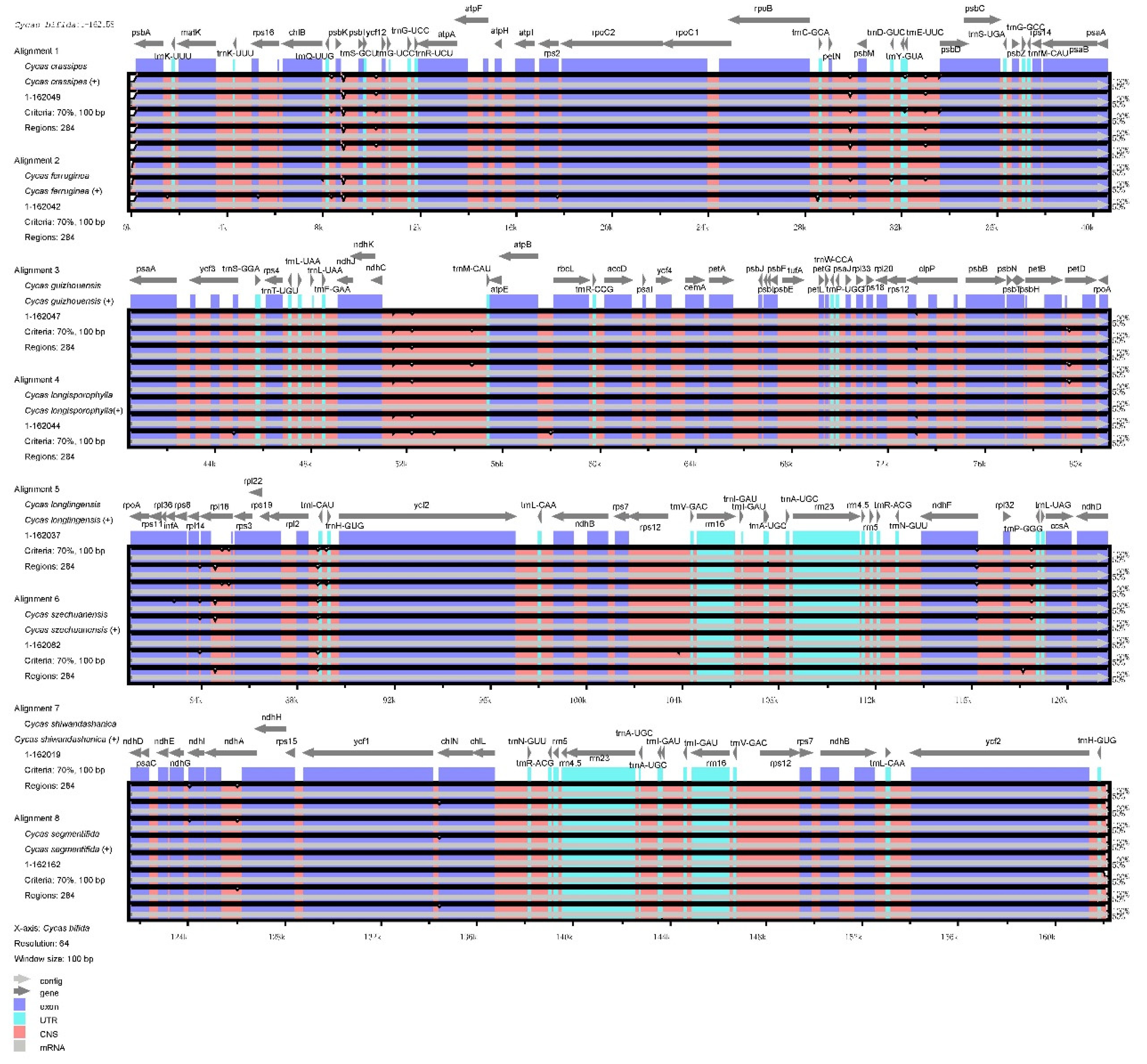
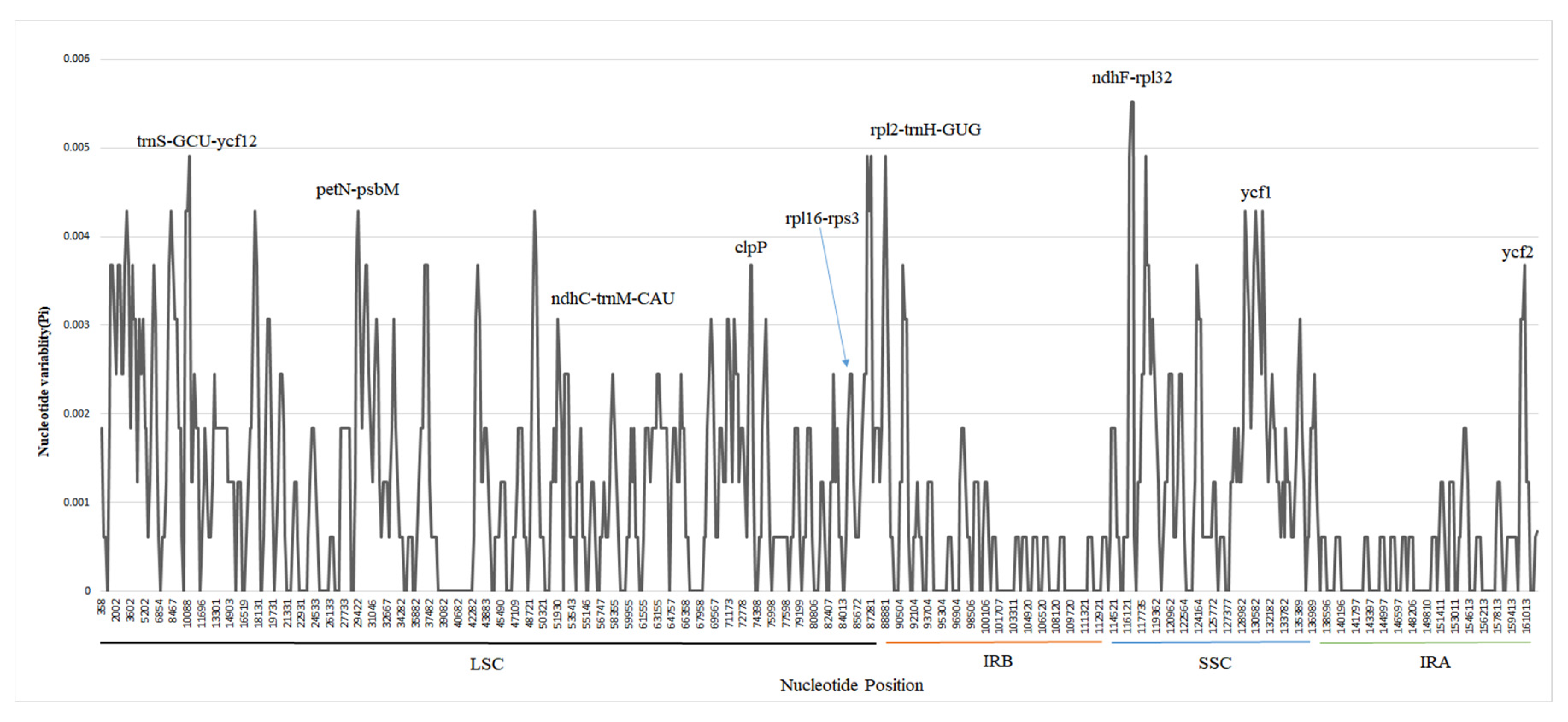
| Sample | Total genome | LSC | IR | SSC | Gene number | |||||||
| Length (bp) | G+C Content (%) |
Length (bp) | G+C Content (%) |
Length (bp) | G+C Content (%) |
Length (bp) | G+C Content (%) | Total | PCGs | tRNA | rRNA | |
| Cycas longisporophylla | 162045 | 39.44 | 88823 | 38.73 | 25049 | 42.03 | 23124 | 36.55 | 131 | 86 | 37 | 8 |
| Cycas bifida | 162159 | 39.42 | 88946 | 38.7 | 25053 | 42.02 | 23107 | 36.52 | 131 | 86 | 37 | 8 |
| Cycas guizhouensis | 162048 | 39.42 | 88826 | 38.71 | 25060 | 42.01 | 23102 | 36.54 | 131 | 86 | 37 | 8 |
| Cycas crassipes | 162050 | 39.42 | 88828 | 38.71 | 25060 | 42.01 | 23102 | 36.54 | 131 | 86 | 37 | 8 |
| Cycas longlingensis | 162038 | 39.44 | 88819 | 38.72 | 25049 | 42.03 | 23121 | 36.55 | 131 | 86 | 37 | 8 |
| Cycas ferruginea | 162043 | 39.43 | 88823 | 38.72 | 25049 | 42.03 | 23122 | 36.55 | 131 | 86 | 37 | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




