1. Introduction
According to the International Agency for Research on Cancer (IARC), colorectal cancer (CRC) is, after lung cancer, the second most common cause of cancer mortality worldwide, with an estimated incidence rate of 935,173 deaths (IRD) of 9.0 per 100,000 people in 2020 [
1]. Incidence rates for CRC increased by 1%-2% annually in young adults (ages <55 years). In the 1990s, CRC was the fourth-leading cause of cancer death in both men and women younger than 50 years, but is now the first in men and the second in women [
2]. In 2020, more than 1.9 million cases of colorectal cancer (CRC) were recorded, and almost 0.9 million patients died of CRC worldwide. There are several possible risk factors that may lead to the development of CRC: Alcohol consumption, smoking, obesity, sedentary lifestyle, unhealthy diet (high intake of red and processed meat and fat) psychological stress, gender, genetic predisposition, family history of CRC, abdominopelvic radiation, personal history of other diseases and intestinal microbiota [
3]. The homeostatic microbiome (microbiota) is the set of all microorganisms that inhabit the human body and that have a key role and influence on other physiological systems. Disruption of the homeostatic microbiome undoubtedly leads to the development of various diseases, as has been demonstrated for impaired insulin secretion and diabetes [
4,
5]. Decreased diversity of the gut microbiome, where one species suppresses another, is closely associated with the development of CRC [
6]. Special analyses found a negative correlation between the Lachnospiraceae species in the gut and the risk of CRC, and a positive correlation between Porphyromonadaceae species, the genus Lachnospiraceae UCG010, the genus Lachnospiraceae, and the genus Selimonas in the gut and CRC risk. These findings suggest a causal relationship between the gut microbiome and the risk of CRC [
7]. The aim of this study was to determine which types of microorganisms in patients with CRC may influence the development and progression of tumor tissue by analyzing the microbial composition of the mucosa on the surface of healthy colon, the surface of tumor tissue, and in the center of tumor tissue.
2. Material and Methods
All patients with colorectal carcinoma were operated on at the Clinic for Digestive Surgery, UKCS, using the open method through medial laparotomy. Depending on the anatomical location of the tumor, and in accordance with oncological principles, right hemicolectomy and extended right hemicolectomy were performed with digestive tract reconstruction, creating a manual ileocolic termino-lateral anastomosis in two layers for carcinomas of the right colon and hepatic flexure. For carcinomas of the left colon and rectum, depending on the anatomical tumor location and disease stage, left hemicolectomy, upper rectal resection, and low rectal resection with stapled colo-rectal termino-terminal anastomosis without protection, as well as colo-anal termino-terminal anastomosis with protective ileostomy, were performed. All subjects enrolled in this research provided informed consent, which was approved by the Ethics Committee on Human Research of UKCS, protocol number 420/25, and this protocol was deemed acceptable by the committee. From 2021, 43 patients were operated on, including 25 men and 19 women, aged between 26 and 87 years (61.11 ± 08). All patients were Caucasian. For research purposes, three swabs were taken: from the healthy mucosa of the colon, from the surface of the tumor tissue, and from the center of the tumor tissue after tumor resection. The patient material samples were cultured on blood agar plates, MacConkey agar, Salmonella-Shigella (SS) agar, Schaedler agar plates, Sabouraud dextrose agar, Schaedler broth, selenite broth, and thioglycolate broth. Solid media were inoculated semi-quantitatively (four-quadrant method) to estimate the number of colonies grown. The inoculated media were incubated under aerobic and anaerobic conditions at 33°C ± 2°C for 48 hours. Enrichment media were subcultured. Identification was performed using standard microbiological methods and the VITEK2 Compact system (bioMérieux, France). Sensitivity testing of isolated strains was conducted using the Kirby-Bauer method (Mueller-Hinton II agar plates [Torlak, Belgrade, Serbia]) and the VITEK-Compact system. The results were interpreted based on EUCAST standards [
8,
9]. Due to the specific nature of the research and the percentage-based presentation of results, statistical tests were not conducted.
3. Results
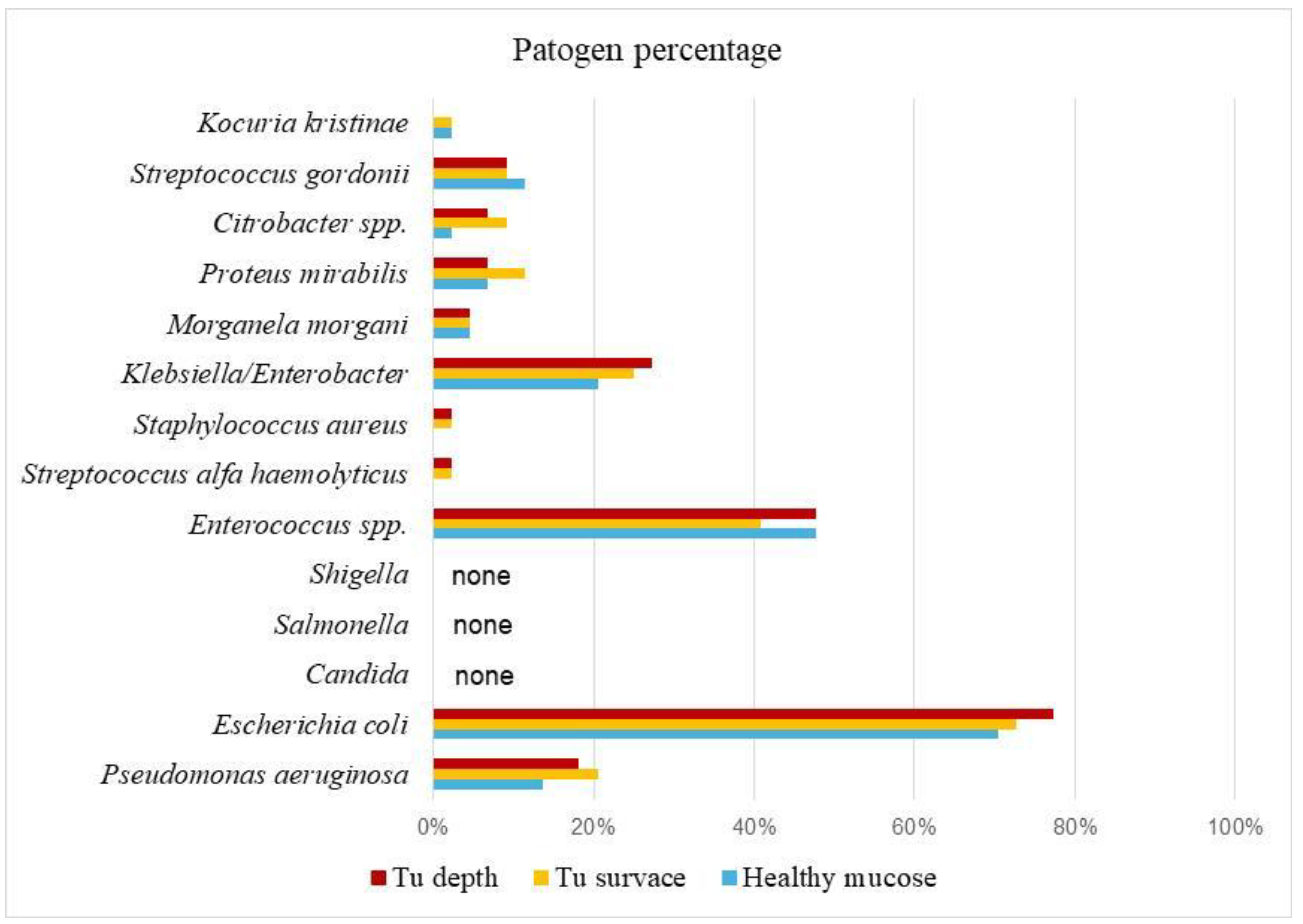
In this study, microbiological analysis data were processed for 132 samples from 44 patients with CRC (colorectal carcinoma), and the results are presented in
Table 1. A total of 15 microorganisms were identified. In the healthy mucosa of the colon, the most prevalent was
E. coli at 70.5%, followed by
Enterococcus spp. (47.7%) and
Klebsiella/Enterobacter (20.5%). Other bacteria were found in smaller percentages:
Pseudomonas aeruginosa 13.6%,
Proteus mirabilis 6.8%,
Streptococcus gordonii 11.4%,
Morganella morganii 4.5%,
Citrobacter spp., and
Kocuria kristinae 2.3% (
Table 1). Microbiological analysis of swabs from the surface of tumor tissue showed that
E. coli was the most prevalent at 72.7%, followed by
Enterococcus spp. 40.9%,
Klebsiella/Enterobacter 25%, and
P. aeruginosa 20%. Other bacteria were present in percentages ranging from 2.3% to 11.4%.
Candida,
Salmonella, and
Shigella were not detected (
Table 1,
Figure 1). In the center of tumor tissue,
E. coli was the most prevalent in 77.3% of samples, followed by
Enterococcus spp. 47.7%,
Klebsiella 27%, and
P. aeruginosa 18.2%. Other bacteria were present in smaller amounts, ranging from 2.3% to 9.1% (
Table 1). By comparing all three types of swabs, it is particularly interesting to note that
E. coli appears in the highest percentage: 70.5% in healthy mucosa, 72.7% on the tumor surface, and 77.3% in the center of tumor tissue.
Enterococcus spp. ranked second with 47.7%, 40.9%, and 47.7%, respectively, followed by
Klebsiella/Enterobacter (20.5%, 25%, 27.3%) and
P. aeruginosa (13.6%, 20.5%, 18.2%).
P. aeruginosa was found in about 13% of samples from healthy mucosa, but its presence increased to up to 20% on the tumor surface and in tumor tissue (
Table 1,
Figure 1).
Candida,
Salmonella, and
Shigella were not found in any of the three types of swabs (healthy mucosa surface, tumor surface, or tumor tissue center). Other bacteria were present in smaller percentages, ranging from 2.3% to 11.4%.
4. Discussion
The analysis of the results of this study identifies four types of bacteria E. coli, Enterococcus spp., Klebsiella/Enterobacter, and Streptococcus gordonii that are present (70-10%) in all three types of samples (from the surface of the healthy intestinal mucosa, the surface of the tumor, and the core of the tumor tissue.
4.1. Escherichia coli
Among these bacteria,
E. coli was the most prevalent, both on the surface of the healthy intestinal mucosa, the tumor surface, and within the tumor tissue, appearing in approximately 70% of cases. In addition to being one of the most common pathogens identified in medical oncology hospitals,
E. coli is also a component of the saprophytic intestinal flora in humans and animals. Its presence is essential for digestion and the synthesis of certain substances, such as vitamin K. However, if the homeostasis of the microbiome is disrupted, an overgrowth of
E. coli can lead to various diseases of the gastrointestinal tract, the urogenital system, the lungs, and in severe cases, sepsis and meningitis. These conditions are most often caused by enteropathogenic, enterotoxic, and enteroinvasive types of
E. coli. Community-acquired
E. coli infections are generally uncommon and can be detected in asymptomatic cancer patients. Cancer has been shown to be a significant risk factor for contracting E. coli infections due to immune deficiencies. Neutropenic enterocolitis is a severe form of diarrhea that should be considered in cancer patients with diarrhea caused by
E. coli. Persistent diarrhea in patients should prompt diagnostic evaluations for colorectal cancer (CRC), as such symptoms may indicate the presence of cancer in the intestinal system [
10]. Recent literature reviews reveal that dysbiosis of the intestinal microbiota is a risk factor for CRC development, with polyketide synthase-positive
Escherichia coli (pks+
E. coli) playing a key role in CRC pathogenesis. Pks+ bacteria produce colibactin, a genotoxic protein that causes DNA damage in host colonocytes. Additionally, these bacteria promote genomic instability, disrupt the intestinal epithelial barrier, induce mucosal inflammation, modulate host immune responses, and influence cell cycle dynamics, creating a microenvironment conducive to tumor initiation and progression [
11]. Biopsy samples analyzed in 1998 using PCR tools detected E. coli in 60% of adenomas and 77% of CRC cases, compared to 12% in adjacent normal biopsies and 3% in normal control samples [
12]. Current studies have shown that the intestinal microbiota, including genera such as
Clostridium,
Bacteroides,
Enterococcus, and
Escherichia, can facilitate colorectal carcinogenesis through direct interaction with host cancer cells, generation of carcinogenic microbial metabolites, and secretion of oncogenic virulence factors [
13,
14,
15,
16].
4.2. Enterococcus spp.
Approximately 40% of the patients analyzed had
Enterococcus bacteria on the surface of the healthy intestinal mucosa, the tumor surface, and in the core of tumor tissue (
Table 1). Enterococci are part of the normal intestinal flora in humans and animals. The most well-known representatives of enterococci are
Enterococcus faecalis and
Enterococcus faecium, which can cause conditions such as endocarditis, urethritis, and sepsis. The connection between
Enterococcus and colorectal cancer is evidenced by findings that co-incubation with conditioned medium of
E. faecalis increased the proliferation of cultured CRC cells. Biliverdin (BV) was identified as a key metabolite produced by
E. faecalis. BV promoted colony formation and cell proliferation while inhibiting cell cycle arrest in cultured CRC cells. BV also significantly increased the expression levels of IL-8 and VEGFA, accelerating angiogenesis in CRC [
17]. Recent studies have shown that the use of auto-probiotics based on non-pathogenic strains of
Enterococcus faecium and
Enterococcus hirae as personalized functional food products (PFFPs) in the complex therapy of early-stage CRC reduced dyspeptic symptoms, postoperative complications, and serum levels of pro-inflammatory cytokines (IL-6 and IL-18) [
18].
4.3. Klebsiella/Enterobacter
In this study, approximately 20% of the patients had this bacterium present in all three samples (on the surface of the healthy intestinal mucosa, the tumor surface, and within tumor tissue).
Klebsiella can be found in water, soil, plants, insects, and is part of the normal flora of humans and animals, colonizing the nose, mouth, and intestines.
Klebsiella species are opportunistic pathogens that can cause pneumonia, urinary tract infections, sepsis, meningitis, diarrhea, peritonitis, and soft tissue infections [
19,
20].
Klebsiella/Enterobacter is among the first intestinal colonizers in newborns. It was established that isolates of
Klebiella pneumonia (51-5) from the intestine of premature children can in ApcMin/+; Il 10−/− mice to induce increased expression of proinflammatory genes with intestinal inflammation. Gnotobiotic experiments in Il10−/− mice have shown that a neonatal isolate induces intestinal inflammation in vivo, with increased expression of proinflammatory genes. Regulation of microbiota composition revealed that
K. pneumoniae 51-5 accelerates the onset of inflammation in Il10−/− mice. Studies have shown that isolates of
K. pneumoniae can induce intestinal inflammation and DNA damage, promoting tumor genesis [
21].
4.4. Streptococcus gordonii
Streptococcus gordonii was detected in approximately 10% of patients across all three sample types.
S. gordonii is a gram-positive bacterium commonly found in the oral cavity as part of the saprophytic flora.
S. gordonii together with related organisms constitute a high percentage, up to 70%, of the bacterial biofilm that forms on clean tooth surfaces. As part of the saprophytic flora, it is harmless in the mouth, however, S. gordonii can cause acute bacterial endocarditis when systemically acquired [
22]. Regarding the connection between CRC and the mentioned bacteria
S. gordonii, there is one case reported in the literature of a patient with endocarditis and an advanced stage of CRC[
23]. In our patients, no other systemic diseases were observed, but it is certain that S.gordoni has an influence on the development of CRC. It has been reported in the literature that
S. gordonii, which participates in polybacterial infection of the gingiva causing periodontitis, also causes an increase in miRNA. Certain types of miRNA have also been reported in various forms of systemic diseases such as atherosclerosis, osteoarthritis, diabetes, obesity, and several cancers [
24].
4.5. Pseudomonas aeruginosa
About 13% of
Pseudomonas is found in the intestinal mucosa, however, its presence increases on the surface of the tumor and in the tumor tissue up to 20%.
Pseudomonas aeruginosa is a clinical bacterium that favors moist environments and is often the cause of various systemic infections in hospitalized patients who are under long-term antibiotic or immunosuppressive therapy. There are data that
P. aeruginosa in human melanoma cell culture causes apoptosis of cancer cells [
25].
P. aeruginosa produces one type of protein, Azurin, which negatively affects the growth and development of cancer cells in both human melanoma and breast cancer cells [
26,
27]. That the relationship between the aforementioned bacteria and cancer cells is very complex, is shown by the data that cancer cells secrete aldolase, which increases the adhesion of
Bacillus pseudomonas to the surface of cancer cells [
27]. Examining the routes and types of infection in patients with adenocarcinoma of the pancreas, the presence of
P. aeruginosa in the tumor tissue was determined in 33% of the subjects [
28]. The fact that
P. aeruginosa in pancreatic islet culture significantly reduces insulin secretion shows a very complex relationship between the mentioned bacteria and certain types of human host cell [
29]. In addition to the fact that
Pseudomonas has a very harmful effect and is a frequent cause of pneumonia and sinusitis in immunocompromised patients as well as complications in puncture wounds and burns, its presence in certain types of cancer obviously has a suppressive effect on the growth of cancer cells. So far, no data have been presented in the literature that indicate the existence and association of
P. aeruginosa and colorectal cancer cells, so we can only assume that the presence of
Pseudomonas can be favorable for CRC patients and negatively affect the development of CRC cell.
5. Conclusions
There is a very complex relationship between the microorganisms of the homeostatic microbiota in a healthy person, and whose disruption due to the action of various factors can lead to the development of conditional pathogens and the development of various diseases, in this case CRC. Certain bacteria favor the occurrence and development of CRC, while some can suppress the growth of cancer cells. Regular analyses of the microbiological composition of the human stool can be a prevention for the occurrence of CRC as well as a referral for more sensitive diagnostic methods. Due to the competitive relationship of microorganisms of the saprophytic intestinal flora, in the future we should seriously deal how to suppress the growth and overgrowth of pathogens that are associated with the occurrence and development of CRC.
Author Contributions
The following statements should be used “Conceptualization, D.N ; Methodology, S.L and S.J.; Software, I.S.; Validation, D.N., J.J., I.S., D.G., S.J., S.L. and V.S..; Formal Analysis, D.N..; Investigation, D.N., V.S. and J.J..; Resources, S.L..; Data Curation, D.N., I.S..; Writing – Original Draft Preparation, D.N.; Writing – Review & Editing, D.N.; Visualization, D.N.; Supervision, D.N.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of University Klinical Center of Serbia, protocol code 420/25, date of approval: 26.12.2024.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
To exclude this statement.
Conflicts of Interest
The authors declare no conflict of interest.
Clinical trials : cross-sectional studies.
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Available online: https://gco.iarc.fr/today (accessed on 10 October 2023).
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer Statistics, 2024. CA: A Cancer Journal for Clinicians 2024, 74. [Google Scholar] [CrossRef] [PubMed]
- Roshandel, G.; Ghasemi-Kebria, F.; Malekzadeh, R. Colorectal Cancer: Epidemiology, Risk Factors, and Prevention. Cancers 2024, 16, 1530. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, D. Diabetes Mellitus and Obesity as a Result of a Disrupted Homeostatic Microbiome. New Data on Etiopathogenesis of Diabetes Mellitus. Vojnosanitetski pregled 2018, 75, 1110–1117. [Google Scholar] [CrossRef]
- Nikolic, D.M.; Dimitrijevic-Sreckovic, V.; Ranin, L.T.; Stojanovic, M.M.; Ilic, I.D.; Gostiljac, D.M.; Soldatovic, I.A. Homeostatic Microbiome Disruption as a Cause of Insulin Secretion Disorders. Candida albicans, a New Factor in Pathogenesis of Diabetes: A STROBE Compliant Cross-Sectional Study. Medicine 2022, 101, e31291. [Google Scholar] [CrossRef] [PubMed]
- Rebersek, M. Gut Microbiome and Its Role in Colorectal Cancer. BMC Cancer 2021, 21. [Google Scholar] [CrossRef]
- Ma, M.; Zheng, Z.; Li, J.; He, Y.; Kang, W.; Ye, X. Association between the Gut Microbiota, Inflammatory Factors, and Colorectal Cancer: Evidence from Mendelian Randomization Analysis. Frontiers in Microbiology 2024, 15. [Google Scholar] [CrossRef] [PubMed]
- Ratnesh, K.; Jha, S.; Arya, A. Clinical and Bacteriological Profile of Abdominal Surgical Site Infections in an Indian Hospital. Bioinformation 2022, 18, 962–967. [Google Scholar] [CrossRef] [PubMed]
- Rasilainen, S.K.; Juhani, M.P.; Kalevi, L.A. Microbial Colonization of Open Abdomen in Critically Ill Surgical Patients. World Journal of Emergency Surgery 2015, 10. [Google Scholar] [CrossRef]
- Joob, B.; Wiwanitkit, V. Cancerous Patients and Outbreak of Escherichia coli: An Important Issue in Oncology. Asian Pacific Journal of Tropical Disease 2014, 4, 204–206. [Google Scholar] [CrossRef]
- Sadeghi, M.; Mestivier, D.; Sobhani, I. Contribution of Pks+ Escherichia coli (E. coli) to Colon Carcinogenesis. Microorganisms 2024, 12, 1111–1111. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Khilkin, M.; Kerjaschki, D.; Schreiber, S.; Ortner, M.; Weber, J.; Lochs, H. Association between Intraepithelial Escherichia coli and Colorectal Cancer. Gastroenterology 1998, 115, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Yu, J. Gut Microbiota in Colorectal Cancer: Mechanisms of Action and Clinical Applications. Nature Reviews Gastroenterology & Hepatology 2019, 16. [Google Scholar] [CrossRef]
- Humphries, J.D. Integrin Ligands at a Glance. Journal of Cell Science 2006, 119, 3901–3903. [Google Scholar] [CrossRef]
- Rubinstein, M.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Yiping W. Fusobacterium Nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via Its FadA Adhesin. Cell Host & Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef]
- Louis, P.; Hold, G.L.; Flint, H.J. The Gut Microbiota, Bacterial Metabolites and Colorectal Cancer. Nature reviews. Microbiology 2014, 12, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, J.; Deng, M.; Chen, X.; Jiang, L.; Zhang, J.; Tao, L.; Yu, W.; Qiu, Y. Enterococcus faecalis Promotes the Progression of Colorectal Cancer via Its Metabolite: Biliverdin. Journal of Translational Medicine 2023, 21. [Google Scholar] [CrossRef] [PubMed]
- Ermolenko, E.; Baryshnikova, N.; Alekhina, G.; Zakharenko, A.; Ten, O.; Kashchenko, V.; Novikova, N.; Gushchina, O.; Ovchinnikov, T.; Morozova, A.; Ilina, A.; Karaseva, A.; Tsapieva, A.; Gladyshev, N.; Dmitriev, A.; Suvorov, A. Autoprobiotics in the Treatment of Patients with Colorectal Cancer in the Early Postoperative Period. Microorganisms 2024, 12, 980. [Google Scholar] [CrossRef]
- Bagley, S.T. Habitat Association of Klebsiella Species. Infection control : IC 1985, 6, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Ristuccia, P.A.; Cunha, B.A. Klebsiella. Infection Control 1984, 5, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Pope, J.L.; Yang, Y.; Newsome, R.C.; Sun, W.; Sun, X.; Ukhanova, M.; Neu, J.; Issa, J.-P.; Mai, V.; Jobin, C. Microbial Colonization Coordinates the Pathogenesis of a Klebsiella pneumoniae Infant Isolate. Scientific Reports 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Salvà-Serra, F.; Jakobsson, H.E.; Thorell, K.; Gonzales-Siles, L.; Hallbäck, E.T.; Jaén-Luchoro, D.; Boulund, F.; Sikora, P.; Karlsson, R.; Svensson, L.; Bennasar, A.; Engstrand, L.; Kristiansson, E.; Moore, E.W. Draft Genome Sequence of Streptococcus gordonii Type Strain CCUG 33482 T. Genome Announcements 2016, 4. [Google Scholar] [CrossRef]
- Callejo-Goena, A.; Rubio-Etxebarria, I.; Sancho-Gutiérrez, A.; Azkuna-Sagarduy, J.; Lopetegi-Aizpurua, A.; López-Vivanco, G. Infective Endocarditis in a Patient with Metastatic Colorectal Cancer. Revista Espanola de Quimioterapia 2018, 31, 75–77. [Google Scholar] [PubMed]
- Aravindraja, C.; Jeepipalli, S.; Duncan, W.D.; Vekariya, K.M.; Rahaman, S.O.; Chan, E.K.L.; Kesavalu, L. Streptococcus gordonii Supragingival Bacterium Oral Infection-Induced Periodontitis and Robust MiRNA Expression Kinetics. International Journal of Molecular Sciences 2024, 25, 6217–6217. [Google Scholar] [CrossRef]
- Yamada, T.; Goto, M.; Punj, V.; Zaborina, O.; Kimbara, K.; Das Gupta, T.K.; Chakrabarty, A.M. The Bacterial Redox Protein Azurin Induces Apoptosis in J774 Macrophages through Complex Formation and Stabilization of the Tumor Suppressor Protein P53. Infection and Immunity 2002, 70, 7054–7062. [Google Scholar] [CrossRef] [PubMed]
- Punj, V.; Bhattacharyya, S.; Saint-Dic, D.; Vasu, C.; Cunningham, E.A.; Graves, J.; Yamada, T.; Constantinou, A.I.; Christov, K.; White, B.; Li, G.; Majumdar, D.; Chakrabarty, A.M.; Das Gupta, T.K. Bacterial Cupredoxin Azurin as an Inducer of Apoptosis and Regression in Human Breast Cancer. Oncogene 2004, 23, 2367–2378. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.K.; Naffouje, S.A.; Goto, M.; Wang, J.; Christov, K.; Rademacher, D.J.; Green, A.; Stecenko, A.A.; Chakrabarty, A.M.; Das Gupta, T.K.; Yamada, T. Cross-Talk between Cancer and Pseudomonas aeruginosa Mediates Tumor Suppression. Communications Biology 2023, 6. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, D.; Latincic, S.; Stojanovic, M.; Grubor, N.; Ranin, L.; Nation, B. Routes and Types of Microbial Infection in the Pathology of Pancreatic Adenocarcinoma. Srpski arhiv za celokupno lekarstvo 2021, 149, (9–10). [Google Scholar] [CrossRef]
- Nikolić, D.M. Effects of Bacterial Infection on Insulin Secretory Capacity of Human Adult Pancreatic Islets. British Journal of Biomedical Science 2011, 68, 181–184. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).