1. Introduction
1.1. Ion Channels
Ion channels, as the name suggests, as we deconstruct the terminology, in a lay man’s term means, a channel or a passage that helps pass the ions. When we talk of passage, then the flow of the ions has to happen through something permeable, a wall, a barrier, an obstruction, etc. Also, the channel should open and close with some purpose and there has to be some definite aim, for which the channel behind which the passage will work. For other conditions, the channel has to remain closed. Investigating these conditions, will help in understanding the ambinence of the channel itself. Another aspect of particular importance is the fact regarding the kind of ions that will pass through the passage and why such ions need to pass through such a channel. Along with the study of these channels, comes the study of the varied structure of these channels which help the passage of these ions. All these issues gives rise to the field of simulation and modeling of these channels also.
Broadly speaking, ion channel is one of the two classes of an ionophoric proteins, that will let the ions pass downhill along with the electrochemical gradient in the channel without the input of any metabolic activity; the other being ion transporters. Ionophoric proteins are structures that reversibly binds with ions. Ion transporters are proteins that will carry the ions along with them through a permeable wall or barrier or an obstruction against a concentration gradient. This passage of the ions requires openning and closing mechanism in the channel and in electrophysiology, gating is the term used for such a mechanism. It is the process were the channel transforms from a conducting mode where ions flow to a non-conducting mode were ions do not pass through. The rate of closing and openning is referred to as kinetics of gating.
There are two major kinds of ion channels namely, (a) Voltage gated ion channel and (b) Ligand gated ion channel. The former operates via the difference in barrier or membrane voltage potential while the latter works via the binding of ligand to the channel. Also, there are different kinds of ions that go through the membrane. These include chloride, potassium, sodium, calcium and proton, to name a few. Depending on the kind of ions, certain functionalities are characteristics of a respective ion channel under study. But before I delve deeper into the specific area of interest, it is important to ask, what is the purpose of such a channel and why this gating process in helpful? The answer to this question will also address the ambience in which the channel is working or not working and thus build the context for the above posed question.
1.2. Voltage Gated Proton Ion Channel - HVCN1
The voltage gated proton ion channel is a unique channel unlike other ion channels, as it has some special properties. The gated channel is not a pump as it works without the help of metabolic activity of ATP. It is a higly selective channel and has a perfect selectivity in comparison to the other ion channels which are sometime imperfect in nature. With strong dependence on the temperature and the lack of an aqueous pore that probably makes the selectivity extremely perfect, the channel distinguishes in making the flow of the hydrogen proton outward. Also, since the flow of protons can happen in various ways, the route of which the other ions cannot take, voltage gated ion channels form a separate class in themselves.
That, HVCN1 is the gene that encodes for voltage gated proton channel was discovered recently in 2006 by two different group simulataneously (i.e Ramsey et al. [
1] and Sasaki et al. [
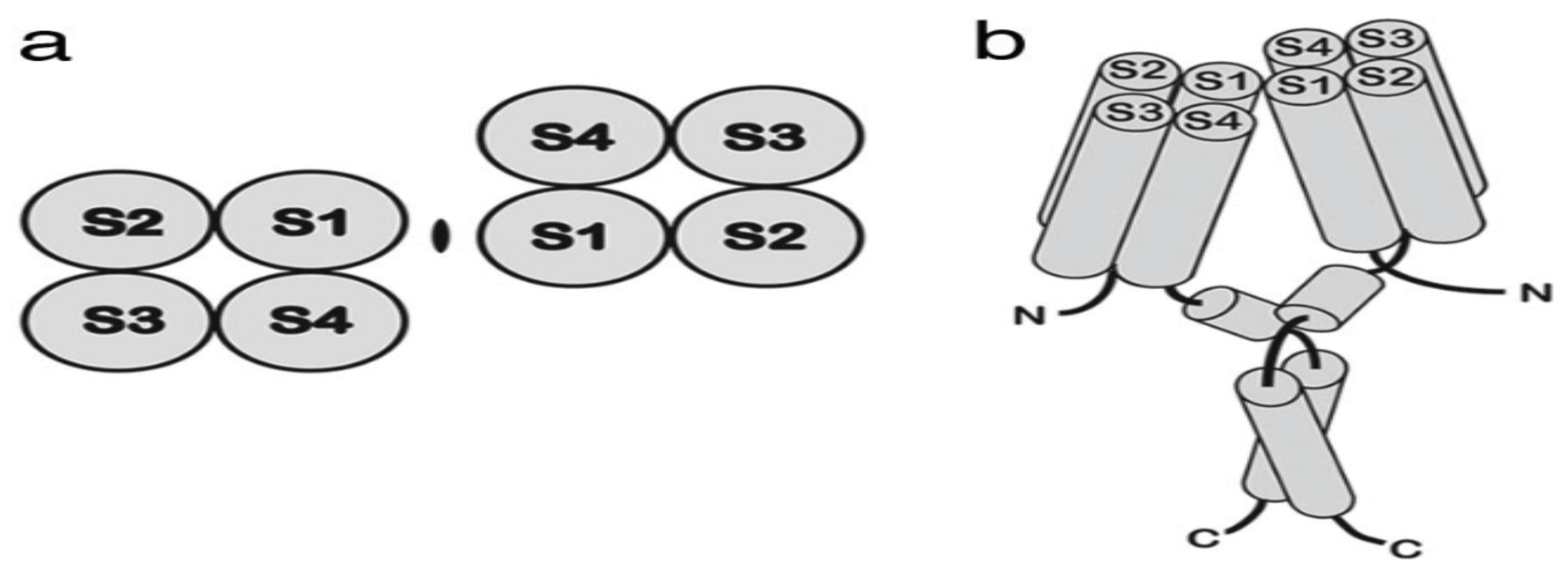
2]). The channels resembles the voltage sensing domain VSD of ordinary voltage gated cation channels. Structurally, the proton channel constitutes four membrane-spanning region S1-S4 and lacks the separate pore domain. The human Hv1 forms a dimer in the membrane. Lee et al. [
3] shows this dimeric structure in
Figure 1 as a probable model. Advanced review on the topic can be found in Rebolledo et al. [
4].
Earlier we talked about the regulation of the . This regulation happens by balancing the acidity, i.e the concentration of the hydrogen protons in the cells. What we find is that, if there is excess of concentration of the hydrogen ions in the cell, then the proton channel works by gating the protons in an outward direction to stabilize the concentration to a physiological optimum for proper functioning of the cell. However, when the protons are being directed outside, the balance of the concentration in the thriving environment of the cell is disturbed. To compensate for this, simultaneously, a flux of the electrons is made possible via NADPH oxidase. Thus the balance in concentration is maintained. As a consequence of the above phenomena, one of the jobs of proton channels is to regulate the acidity by extrusion of the acidic material at intracellular level and thus increase the . Simultaneously, from the extracellular perspective the acid secretion leads to the regulation of acidity outside the cell and hence the decrease of . Finally, the extrusion of protons has a controlled effect on the osmotic swelling in comparison to other protons.
1.3. HVCN1 in Colorectal Cancer
Wang et al. [
5] have recently shown that HVCN1 was found to be highly expressed in colorectal cancer cell lines. Suppression of Hv1 in highly metastatic colorectal cancer restrained the extrusion of protons and thus leading to low
. Since the expression of Hv1 was found to be very high in colorectal cancer cells, the extrusion of protons is expected to be high. What this means is that there is an increase in the
and cancer cells are able to thrive in hypoxic environments where the acidity of the envirnoment is beyond the normal level. A more recent role of voltage-gated proton channel (Hv1) in cancer biology has been studied by Alvear-Arias et al. [
6]. El Chemaly et al. [
7] in their study, show that the genetic deletion of Hv1 delays tumor development in a mouse model of granulocytic sarcoma and report the discovery and characterization of two novel bioavailable inhibitors of Hv1 channels that they validate by orthogonal assays and electrophysiological recordings. HVCN1 was found to be highly suppressed in colorectal cancer cell lines after the treatment of ETC-1922159 drug as observed in Madan et al. [
8]. Most studies till now, have dealt with how the HVCN1 works and there is very less informnation reagarding which gene/protein combinations might be working synergistically along with it. It would be nice to observe if there is any connection between the independently observed factors in the form of unknown biological hypotheses. To solve the issue, the next section addresses poses a solution to the problem.
1.4. Combinatorial Search Problem and a Possible Solution
In a recently published work Sinha [
9], a frame work of a search engine was developed which can rank combinations of factors (genes/proteins) in a signaling pathway. Readers are requested to go through the adaptation of the above mentioned work for gaining deeper insight into the working of the pipeline and its use of published data set generated after administration of ETC-1922159, Sinha [
10]. The work uses SVM package by Joachims [
11] in
https://www.cs.cornell.edu/people/tj/svm_light/svm_rank.html. I use the adaptation to rank 2
nd order gene combinations.
2. Results & Discussion
2.1. HVCN1 Related Synergies
2.1.1. HVCN1-NOX1/ENOX2
The reactive oxygen species (ROS)-producing enzyme NADPH oxidase (NOX) was first identified in the membrane of phagocytic cells. Its discovery and molecular mechanisms has been described in detail in Vermot et al. [
12]. Hv1 channels sustain high-level production of ROS by NOX2 in phagocytes (Ramsey et al. [
13], El Chemaly et al. [
14] and El Chemaly et al. [
7]). In colorectal cancer cells treated with ETC-1922159, NOX1 and HVCN1, were found to be down regulated and recorded independently. Ecto-NOX disulfide-thiol exchanger 2 (ENOX2), is a member of the NOX family of NADPH oxidases. ENOX2 proteins are restricted to cancer (Hostetler et al. [
15], Chueh et al. [
16]). In colorectal cancer cells treated with ETC-1922159, ENOX2 and HVCN1, were found to be down regulated and recorded independently. I was able to rank 2
nd order combination of NOX1/ENOX2 and HVCN1, that were down regulated.
Table 1 shows rankings of these combinations. Followed by this is the unexplored combinatorial hypotheses in
Table 2 generated from analysis of the ranks in
Table 1. The
Table 1 shows rankings of NOX family w.r.t HVCN1. NOX1 - HVCN1 shows low ranking of 349 (laplace) and 855 (linear). ENOX2 - HVCN1 shows low ranking of 1404 (laplace), 601 (linear) and 1503 (rbf). These rankings point to the synergy existing between the two components, which have been down regulated after the drug treatment.
One can also interpret the results of the
Table 1 graphically, with the following influences - • NOX family w.r.t HVCN1 with HVCN1
NOX1; and HVCN1
ENOX2.
2.1.2. HVCN1-MAPK/MMP
Banskota et al. [
17] show that NOX2-derived ROS regulates NOX1, NOX2, and matrix metalloproteinase-7 (MMP7) expression through the mitogen-activated protein kinase (MAPK) pathway, in colon cancer. Based on this experimentally tested phenomena, there might be possible connection between HVCN1 and MMP/MAPK, though the current understanding of how the synergy works might be lacking. In ETC-1922159 treated colorectal cancer cells, MMP11 and MAPK15 were both down regulated and their regulation was recorded individually. I was able to rank 2
nd order combination of MMP11/MAPK15 and HVCN1, that were down regulated.
Table 3 shows rankings of these combinations. Followed by this is the unexplored combinatorial hypotheses in
Table 4 generated from analysis of the ranks in
Table 3. The
Table 3 shows rankings of MMP11/MAPK15 w.r.t HVCN1. MAPK15 - HVCN1 shows low ranking of 1612 (linear) and 60 (rbf). The ranking for MAPK15 points to the possible synergy existing between the two components, which have been down regulated after the drug treatment. However, the ranking of MMP11 does not point to possible synergy with HVCN1 (majority getting high numerical valued rank for down regulation, see table).
One can also interpret the results of the
Table 3 graphically, with the following influences - • MAPK15 w.r.t HVCN1 with HVCN1
MAPK15.
2.1.3. HVCN1- Clinicopathological Parameters TP53/MKI67/GST
In colorectal cancer, while studying Hv1 as a new potential biomarker for diagnosis and prognosis, Wang et al. [
5] reported clinicopathological parameters like TP53, MKI67 and GST. Correlation between Hv1 expression levels in colorectal cancer and clinicopathological parameters, were recorded. In ETC-1922159 treated colorectal cancer cells, TP53, MKI67 and members of GST family were found down regulated and their regulation was recorded individually. I was able to rank 2
nd order combination of TP53/MKI67/GST and HVCN1, that were down regulated.
Table 5 shows rankings of these combinations. Followed by this is the unexplored combinatorial hypotheses in
Table 6 generated from analysis of the ranks in
Table 5. The
Table 5 shows rankings of TP53/MKI67/GST w.r.t HVCN1. TP53 - HVCN1 shows low ranking of 1532 (laplace) and 1640 (rbf). MKI67 - HVCN1 shows low ranking of 184 (linear) and 572 (rbf). MKI67 - HVCN1 shows low ranking of 184 (linear) and 572 (rbf). GSTO2 - HVCN1 shows low ranking of 689 (laplace), 1341 (linear) and 1591 (rbf). GSTM4 - HVCN1 shows low ranking of 741 (laplace) and 593 (rbf).
One can also interpret the results of the
Table 5 graphically, with the following influences - • TP53 w.r.t HVCN1 with HVCN1
TP53; • MKI67 w.r.t HVCN1 with HVCN1
MKI67; and • GST w.r.t HVCN1 with HVCN1
GSTO2; HVCN1
GSTM4.
2.1.4. HVCN1-PLC
Alvear-Arias et al. [
6], state that the effect called ”enhanced gating mode” is a phenomenon that occurs after stimulation with phorbol 12-meristate-13-acetate (PMA). PMA addition to cells, promotes a shift of activation curves of Hv1 channel currents to negative voltages and faster opening kinetics (Bánfi et al. [
18], DeCoursey et al. [
19]). PMA operates by activating the phospholipase C (PLC) that leads to the activation of protein kinase C (PKC). PKC phosphorylates Threonine 29 in the N-terminal of hHv1, leading to the enhancement of its gating Musset et al. [
20]. These experimental findings point to the synergy that might be existing between PLC and HVCN1. In ETC-1922159 treated colorectal cancer cells, PLC family and HVCN1 were found down regulated and their regulation was recorded individually. I was able to rank 2
nd order combination of PLC family and HVCN1, that were down regulated.
Table 7 shows rankings of these combinations. Followed by this is the unexplored combinatorial hypotheses in
Table 8 generated from analysis of the ranks in
Table 7. The
Table 7 shows rankings of PLC family w.r.t HVCN1. PLCH1 - HVCN1 shows low ranking of 494 (laplace), 929 (linear) and 922 (rbf). PLCB4 - HVCN1 shows low ranking of 1279 (laplace), 1248 (linear) and 677 (rbf). PLCB2 - HVCN1 shows low ranking of 988 (linear) and 1501 (rbf).
One can also interpret the results of the
Table 7 graphically, with the following influences - • PLC family w.r.t HVCN1 with HVCN1
PLCH1; HVCN1
PLCB4; and HVCN1
PLCB2..
3. Conclusion
Presented here are a range of multiple synergistic HVCN1 2nd order combinations that were ranked via a machine learning based search engine. Via majority voting across the ranking methods, it was possible to find plausible unexplored synergistic combinations of HVCN1-X that might be prevalent in CRC cells after treatment with ETC-1922159 drug.
Author Contributions
Concept, design, in silico implementation - SS. Analysis and interpretation of results - SS. Manuscript writing - SS. Manuscript revision - SS. Approval of manuscript - SS
Acknowledgments
Special thanks to Mrs. Rita Sinha and Mr. Prabhat Sinha for supporting the author financially, without which this work could not have been made possible.
Conflicts of Interest
There are no conflicts to declare.
Source of Data
Data used in this research work was released in a publication in Madan et al. [
8].
References
- Ramsey, I.S.; Moran, M.M.; Chong, J.A.; Clapham, D.E. A voltage-gated proton-selective channel lacking the pore domain. Nature 2006, 440, 1213–1216. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Takagi, M.; Okamura, Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science 2006, 312, 589–592. [Google Scholar] [CrossRef]
- Lee, S.Y.; Letts, J.A.; MacKinnon, R. Dimeric subunit stoichiometry of the human voltage-dependent proton channel Hv1. Proceedings of the National Academy of Sciences 2008, 105, 7692–7695. [Google Scholar] [CrossRef] [PubMed]
- Rebolledo, S.; Qiu, F.; Peter Larsson, H. Molecular structure and function of Hv1 channels. Wiley Interdisciplinary Reviews: Membrane Transport and Signaling 2012, 1, 763–777. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, X.; Li, Q.; Zhang, S.; Li, S.J. Human voltage-gated proton channel hv1: a new potential biomarker for diagnosis and prognosis of colorectal cancer. PLoS One 2013, 8, e70550. [Google Scholar] [CrossRef]
- Alvear-Arias, J.J.; Pena-Pichicoi, A.; Carrillo, C.; Fernandez, M.; Gonzalez, T.; Garate, J.A.; Gonzalez, C. Role of voltage-gated proton channel (Hv1) in cancer biology. Frontiers in Pharmacology 2023, 14, 1175702. [Google Scholar] [CrossRef] [PubMed]
- El Chemaly, A.; Jaquet, V.; Cambet, Y.; Caillon, A.; Cherpin, O.; Balafa, A.; Krause, K.H.; Demaurex, N. Discovery and validation of new Hv1 proton channel inhibitors with onco-therapeutic potential. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 2023, 1870, 119415. [Google Scholar] [CrossRef] [PubMed]
- Madan, B.; Ke, Z.; Harmston, N.; Ho, S.Y.; Frois, A.; Alam, J.; Jeyaraj, D.A.; Pendharkar, V.; Ghosh, K.; Virshup, I.H.; et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene 2016, 35, 2197. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S. Machine learning ranking of plausible (un) explored synergistic gene combinations using sensitivity indices of time series measurements of Wnt signaling pathway. Integrative Biology 2024, 16, zyae020. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S. Sensitivity analysis based ranking reveals unknown biological hypotheses for down regulated genes in time buffer during administration of PORCN-WNT inhibitor ETC-1922159 in CRC. bioRxiv, 1809. [Google Scholar]
- Joachims, T. Training linear SVMs in linear time. In Proceedings of the Proceedings of the 12th ACM SIGKDD international conference on Knowledge discovery and data mining.; pp. 2006217–226.
- Vermot, A.; Petit-Härtlein, I.; Smith, S.M.; Fieschi, F. NADPH oxidases (NOX): an overview from discovery, molecular mechanisms to physiology and pathology. Antioxidants 2021, 10, 890. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, I.S.; Ruchti, E.; Kaczmarek, J.S.; Clapham, D.E. Hv1 proton channels are required for high-level NADPH oxidase-dependent superoxide production during the phagocyte respiratory burst. Proceedings of the National Academy of Sciences 2009, 106, 7642–7647. [Google Scholar] [CrossRef] [PubMed]
- El Chemaly, A.; Okochi, Y.; Sasaki, M.; Arnaudeau, S.; Okamura, Y.; Demaurex, N. VSOP/Hv1 proton channels sustain calcium entry, neutrophil migration, and superoxide production by limiting cell depolarization and acidification. Journal of Experimental Medicine 2010, 207, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Hostetler, B.; Weston, N.; Kim, C.; Morré, D.M.; Morré, D.J. Cancer site-specific isoforms of ENOX2 (tNOX), a cancer-specific cell surface oxidase. Clinical Proteomics 2009, 5, 46–51. [Google Scholar] [CrossRef]
- Chueh, P.J.; Kim, C.; Cho, N.; Morre, D.M.; Morre, D.J. Molecular cloning and characterization of a tumor-associated, growth-related, and time-keeping hydroquinone (NADH) oxidase (tNOX) of the HeLa cell surface. Biochemistry 2002, 41, 3732–3741. [Google Scholar] [CrossRef] [PubMed]
- Banskota, S.; Regmi, S.C.; Kim, J.A. NOX1 to NOX2 switch deactivates AMPK and induces invasive phenotype in colon cancer cells through overexpression of MMP-7. Molecular cancer 2015, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bánfi, B.; Schrenzel, J.; Nüsse, O.; Lew, D.P.; Ligeti, E.; Krause, K.H.; Demaurex, N. A novel H+ conductance in eosinophils: unique characteristics and absence in chronic granulomatous disease. The Journal of experimental medicine 1999, 190, 183–194. [Google Scholar] [CrossRef] [PubMed]
- DeCoursey, T.; Cherny, V.; Zhou, W.; Thomas, L. Simultaneous activation of NADPH oxidase-related proton and electron currents in human neutrophils. Proceedings of the National Academy of Sciences 2000, 97, 6885–6889. [Google Scholar] [CrossRef] [PubMed]
- Musset, B.; Capasso, M.; Cherny, V.V.; Morgan, D.; Bhamrah, M.; Dyer, M.J.; DeCoursey, T.E. Identification of Thr29 as a critical phosphorylation site that activates the human proton channel Hvcn1 in leukocytes. Journal of Biological Chemistry 2010, 285, 5117–5121. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).