Submitted:
02 June 2025
Posted:
04 June 2025
You are already at the latest version
Abstract
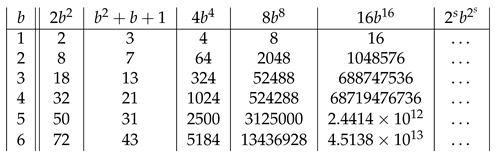
Assembly theory bridges the gap between evolutionary biology and physics by providing a framework to quantify the generation and selection of novelty in biological systems. We formalize the assembly space as an acyclic digraph of strings with 2-in-regular assembly steps vertices and provide a novel definition of the assembly index. In particular, we show that the upper bound of the assembly index depends quantitatively on the number b of unit-length strings, and the longest length N of a string that has the assembly index of N − k is given by N(N−1) = b2 + b + 1 and by N(N−k) = b2 + b + 2k for 2 ≤ k ≤ 9. We also provide particular forms of such maximum assembly index strings. For k = 1, such odd-length strings are nearly balanced. We also show that each k copies of an n-plet contained in a string decrease its assembly index at least by k(n − 1) − a, where a is the assembly index of this n-plet. We show that the minimum assembly depth satisfies d min(N) = ⌈log2(N)⌉, for all b, and is the assembly depth of a maximum assembly index string. We also provide the general formula for the lengths of the minimum assembly index strings having only one independent assembly step in their assembly spaces. Since these results are also valid for b = 1, assembly theory subsumes information theory.
Keywords:
1. Introduction
2. Rudiments
- k copies of a doublet in a string decrease the ASI of this string at least by ;
- k copies of a triplet in a string decrease the ASI of this string at least by ;
- k copies of a minimum ASI quadruplet in a string decrease the ASI of this string at least by ;
- k copies of a maximum ASI quadruplet in a string decrease the ASI of this string at least by ;
- where, the phrase "at least" is meant to indicate that other repetitions, such as e.g. doublets forming multiple quadruplets, etc. can further decrease the ASI of the string. This observation allows us to state the following theorem.
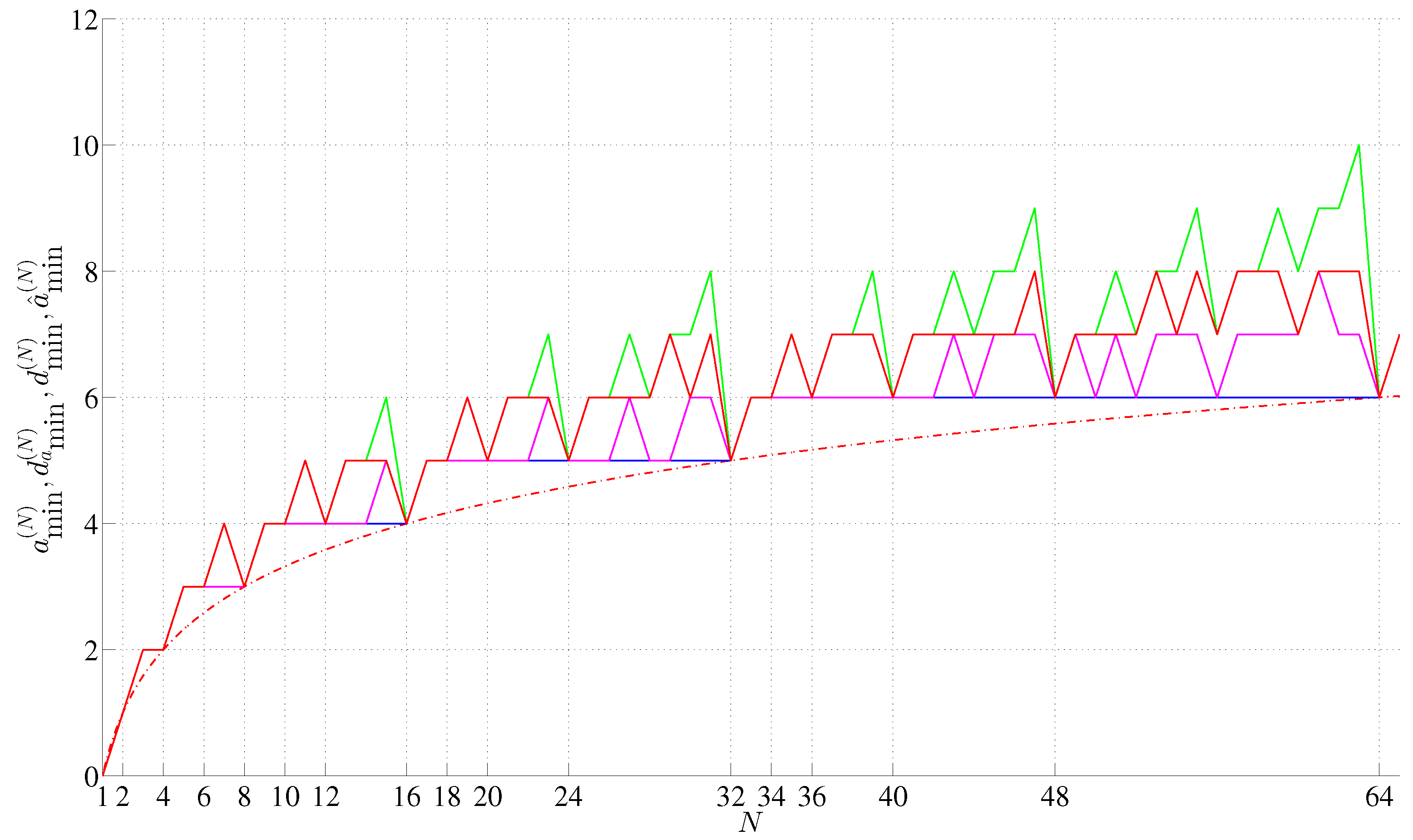
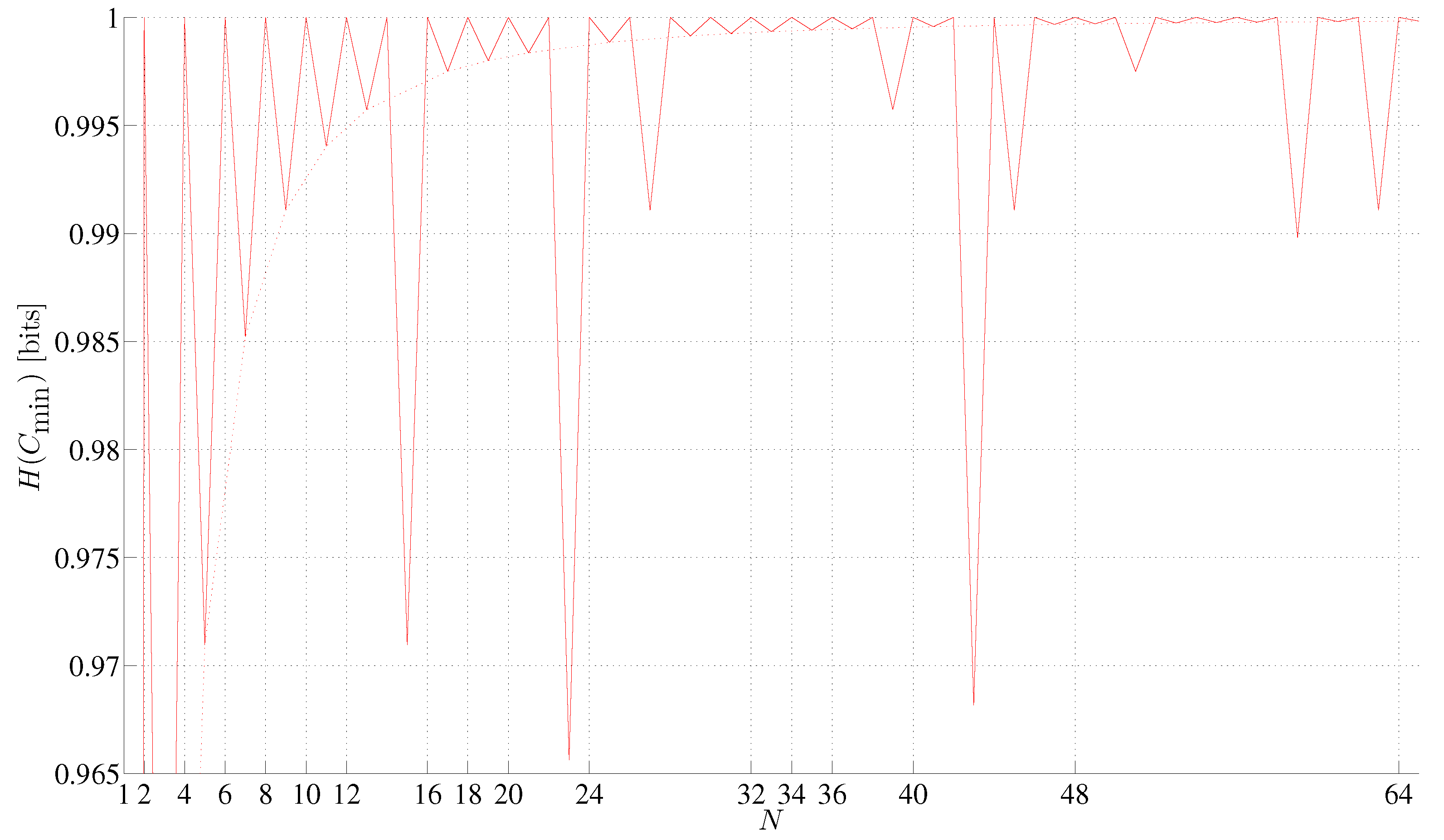
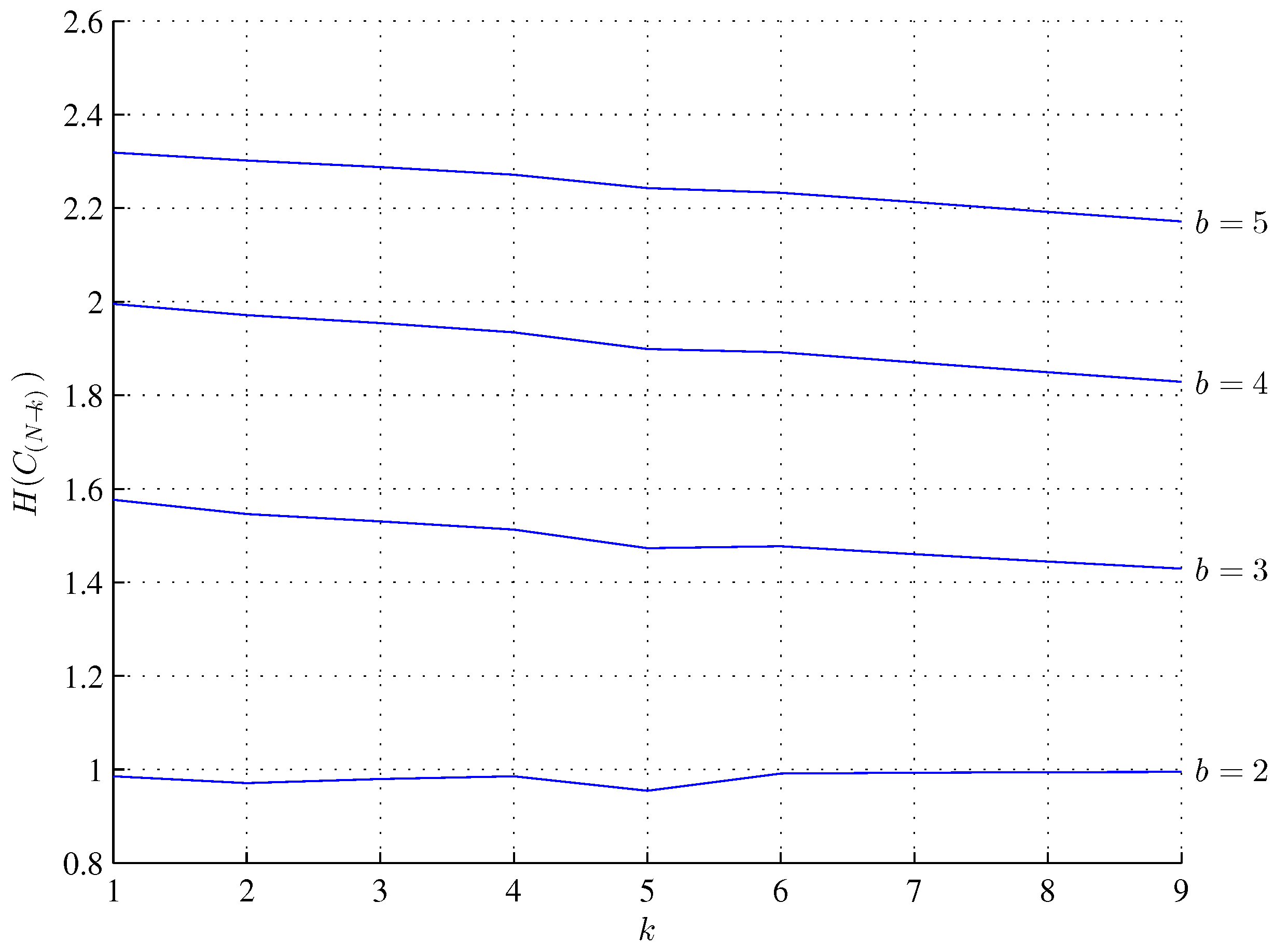
3. Minimum Assembly Depth, Assembly Depth and Entropy of a Minimum Assembly Index, Minimum Assembly Index, and Depth Index
| s | ... | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 2 | 4 | 3 | 7 | 14 | 28 | 56 | 112 | 224 | ... | |
| 15 | 30 | 60 | 120 | 240 | 480 | ||||||
| 23 | 46 | 92 | 184 | 368 | |||||||
| 3 | 2 | 4 | 8 | 3 | 11 | 22 | 44 | 88 | 176 | ... | |
| 27 | 54 | 108 | 216 | 432 | |||||||
| 43 | 86 | 172 | 344 | ||||||||
| 2 | 4 | 8 | 5 | 13 | 26 | 52 | 104 | 208 | ... | ||
| 45 | 90 | 180 | 360 | ... | |||||||
| 4 | 2 | 4 | 8 | 16 | 3 | 19 | 38 | 76 | 152 | ... | |
| 51 | 102 | 204 | 408 | ||||||||
| 83 | 166 | 332 | |||||||||
| 2 | 4 | 8 | 16 | 5 | 21 | 42 | 84 | 168 | ... | ||
| 85 | 170 | 340 | ... | ||||||||
| 2 | 4 | 8 | 16 | 9 | 25 | 50 | 100 | 200 | ... | ||
| 5 | 2 | 4 | 8 | 16 | 32 | 3 | 35 | 70 | 140 | ... | |
| 99 | 198 | 396 | |||||||||
| 163 | 326 | ||||||||||
| 2 | 4 | 8 | 16 | 32 | 5 | 37 | 74 | 148 | ... | ||
| 165 | 330 | ... | |||||||||
| 2 | 4 | 8 | 16 | 32 | 9 | 41 | 82 | 164 | ... | ||
| 2 | 4 | 8 | 16 | 32 | 17 | 49 | 98 | 196 | ... | ||
| 6 | 2 | 4 | 8 | 16 | 32 | 64 | 3 | 67 | 134 | ... | |
| 195 | 390 | ||||||||||
| 323 | |||||||||||
| 2 | 4 | 8 | 16 | 32 | 64 | 5 | 69 | 138 | 276 | ||
| 325 | 650 | ||||||||||
| 2 | 4 | 8 | 16 | 32 | 64 | 9 | 73 | 146 | ... | ||
| 2 | 4 | 8 | 16 | 32 | 64 | 17 | 81 | 162 | ... | ||
| 2 | 4 | 8 | 16 | 32 | 64 | 33 | 97 | 194 | ... | ||
| 7 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 3 | 131 | ... | |
| 387 | |||||||||||
| 2 | 4 | 8 | 16 | 32 | 64 | 128 | 5 | 133 | 266 | ||
| 645 | |||||||||||
| 2 | 4 | 8 | 16 | 32 | 64 | 128 | 9 | 137 | ... | ||
| 2 | 4 | 8 | 16 | 32 | 64 | 128 | 17 | 145 | ... | ||
| 2 | 4 | 8 | 16 | 32 | 64 | 128 | 33 | 161 | ... | ||
| 2 | 4 | 8 | 16 | 32 | 64 | 128 | 65 | 193 | ... |
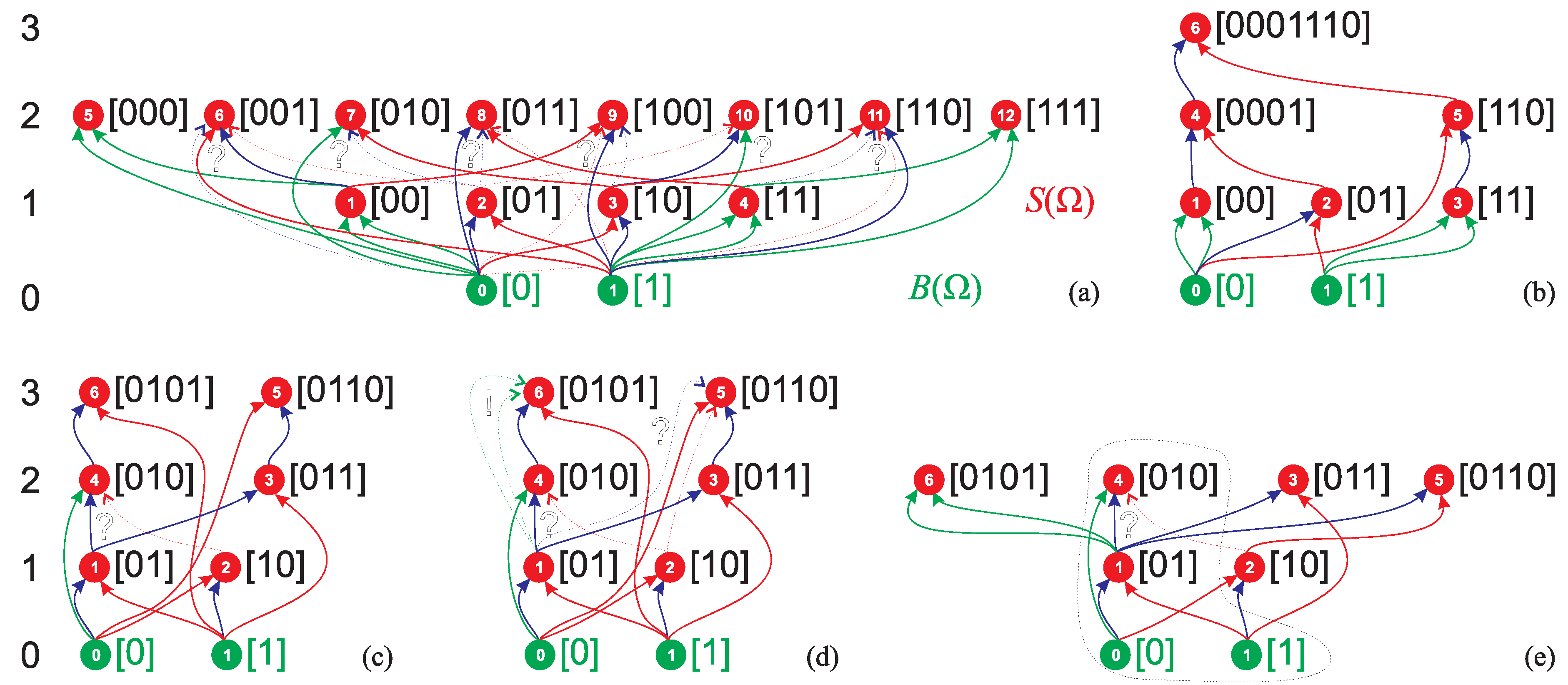
- the of minimum ASI strings having ASI equal to DPI cannot contain strings assembled in independent assembly steps,
- the s of other minimum ASI strings can contain at least two such strings, and therefore
- the assembly space of a maximum ASI string will tend to maximize the number of strings assembled in independent assembly steps in the , taking into account the saturation of the as it cannot contain more than distinct n-plets, and hence to minimize the possible ASD.
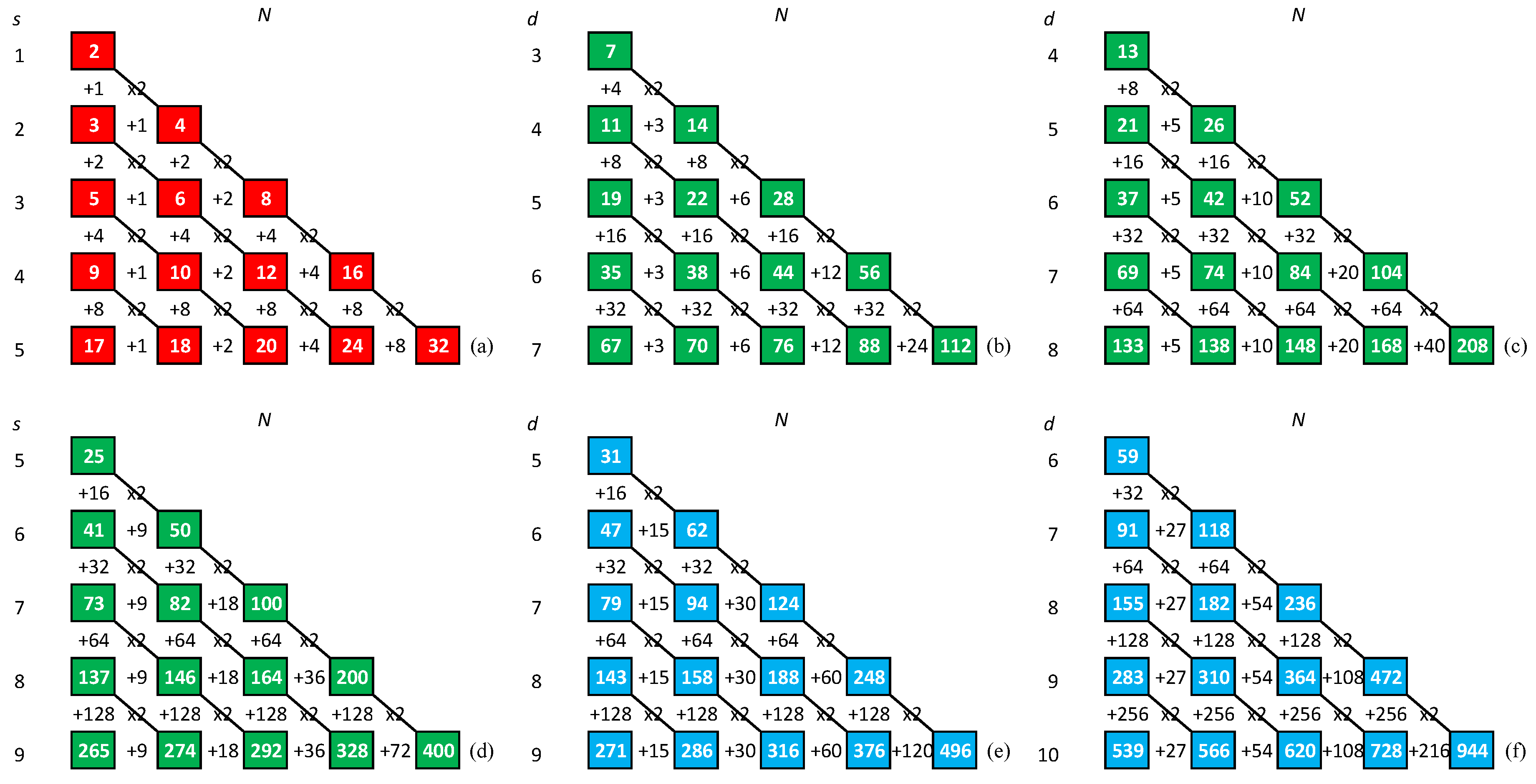
- We note that the difference between the DPI and minimum ASI is, in general, larger than one. The pathways of the minimum ASI strings maximizing the number of independent assembly steps are listed in Table A1 for .
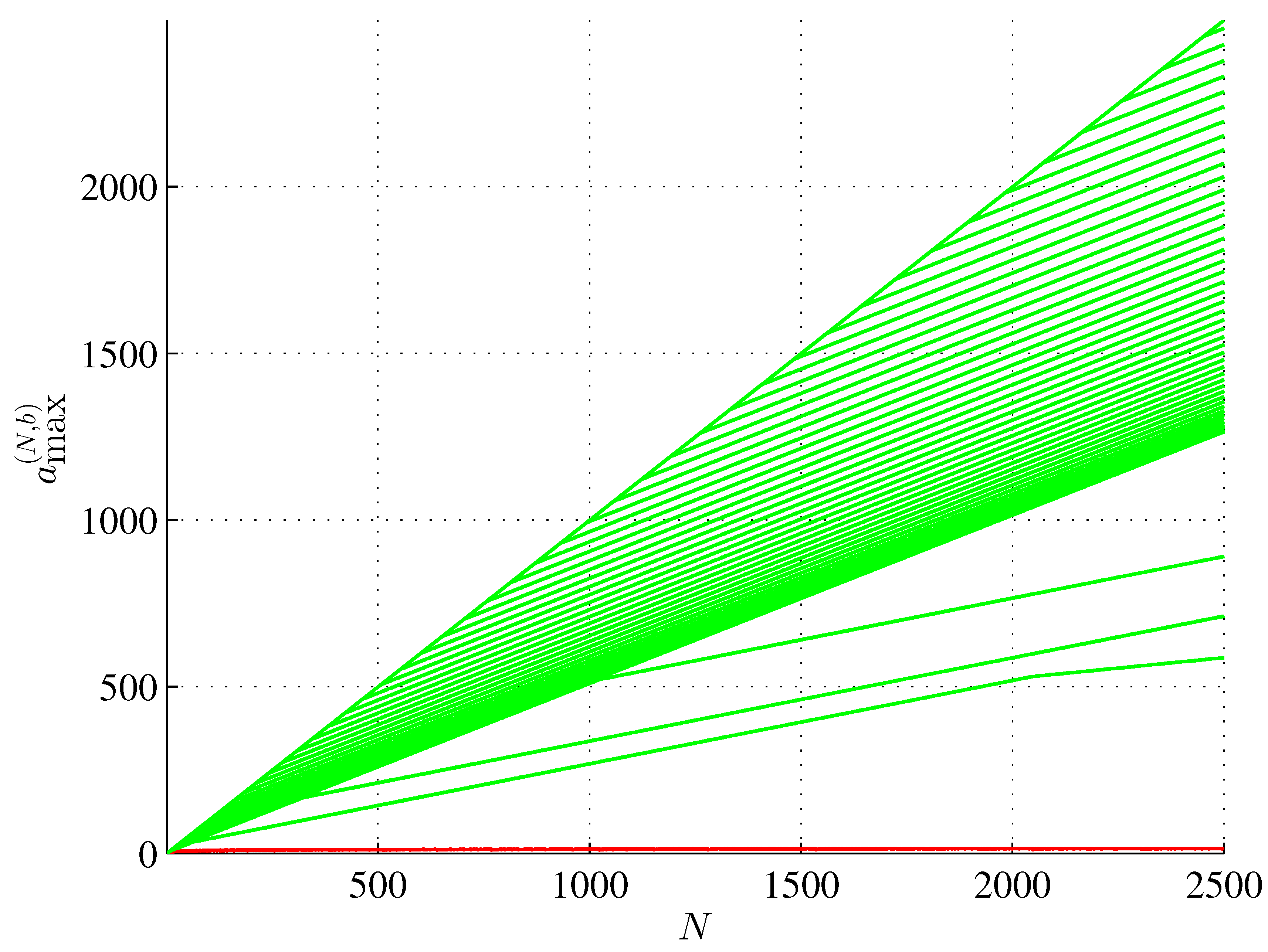
4. Maximum Assembly Index Strings
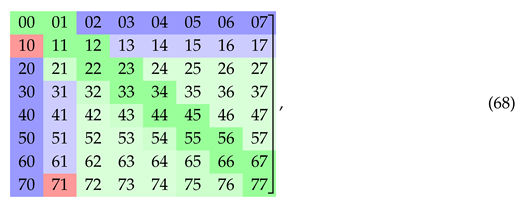
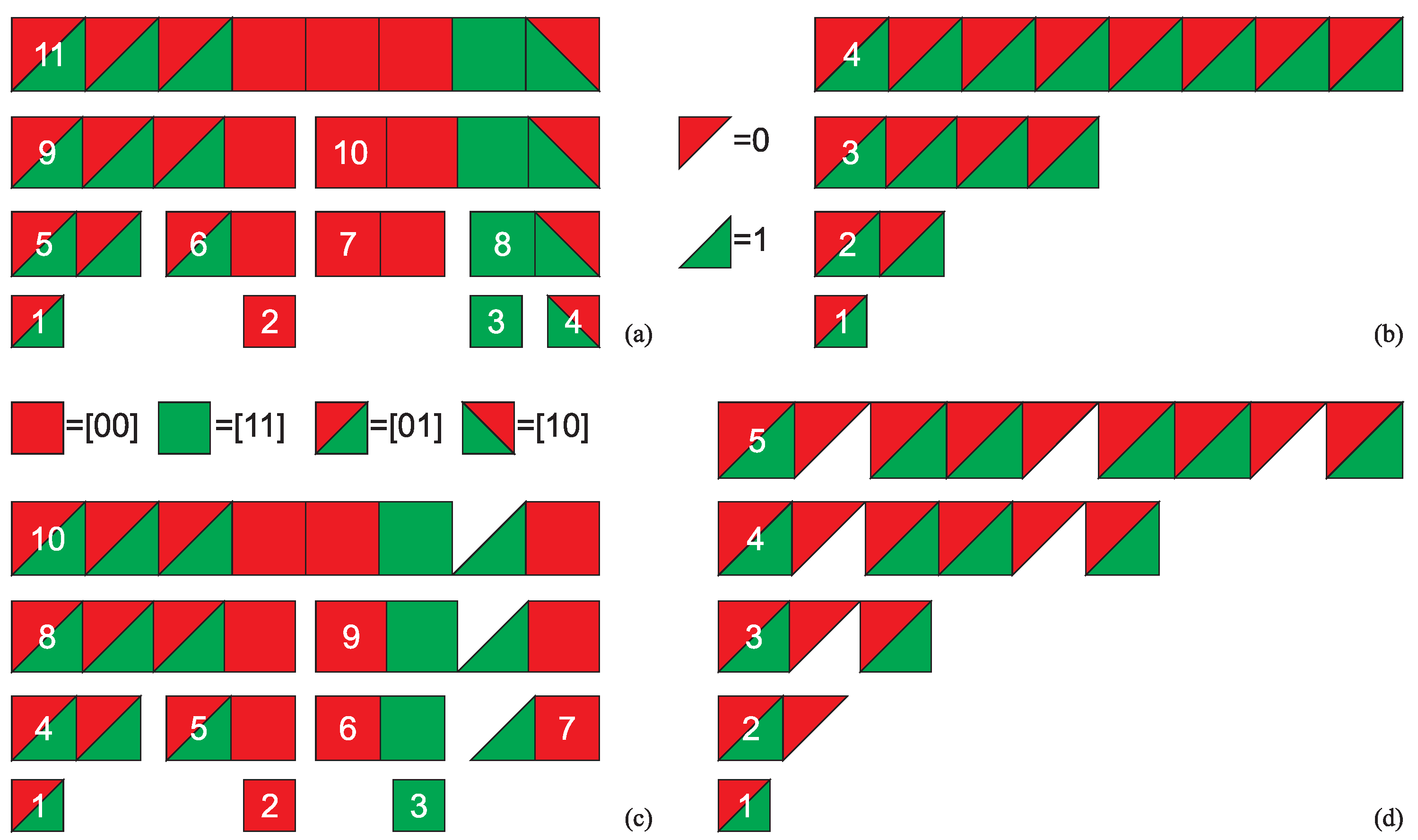
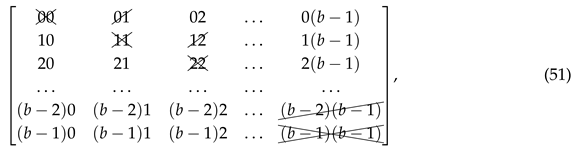 where the crossed out entries on a diagonal cannot be reused, as they would form repetitions in this string. Due to the order of triplets in the string (50) we can also cross out the entries in the first superdiagonal of the matrix (51). By construction, the starting string (50) has length , contains doublets and (if read from left to right) and does not contain doublets , where , , and .
where the crossed out entries on a diagonal cannot be reused, as they would form repetitions in this string. Due to the order of triplets in the string (50) we can also cross out the entries in the first superdiagonal of the matrix (51). By construction, the starting string (50) has length , contains doublets and (if read from left to right) and does not contain doublets , where , , and .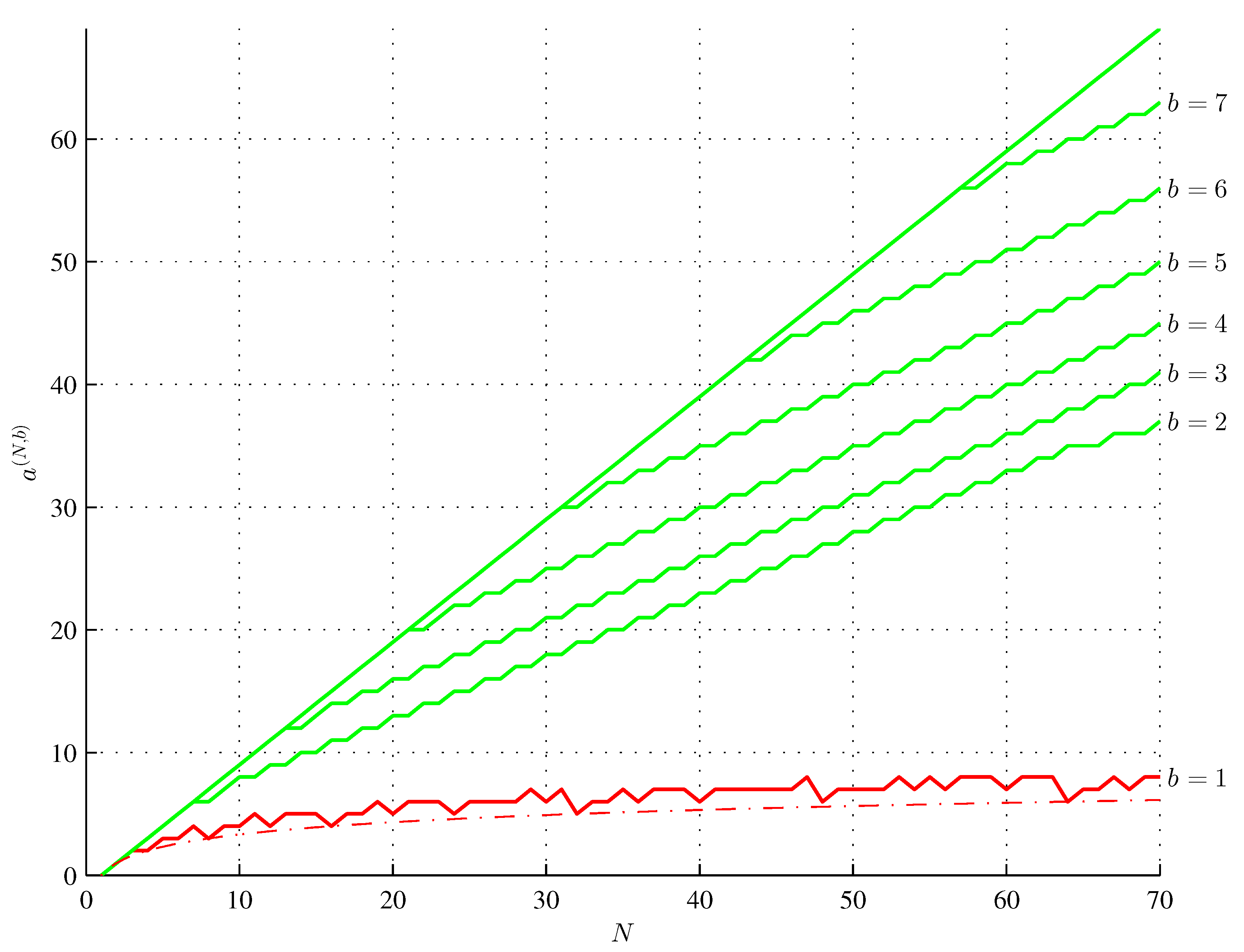
 as
as5. Assembly Index and the Expected Waiting Time
6. A Method of Generating a Maximum Assembly Index String


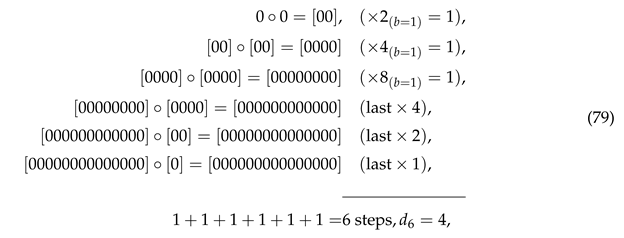 where obviously . However, this is the exception for as the ASI of this string is five if it is assembled using doublet and triplet .
where obviously . However, this is the exception for as the ASI of this string is five if it is assembled using doublet and triplet .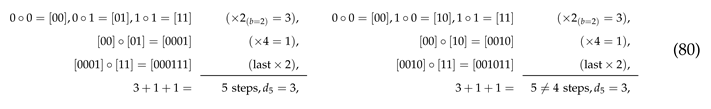 as the right one can be assembled in four steps with . Similarly, only the top distribution of doublets below correctly reflects the maximum ASI of the assembled string for
as the right one can be assembled in four steps with . Similarly, only the top distribution of doublets below correctly reflects the maximum ASI of the assembled string for
 as the bottom one can be assembled in six steps with . Furthermore, this method tends to exaggerate the estimated maximum ASI value, that is,
as the bottom one can be assembled in six steps with . Furthermore, this method tends to exaggerate the estimated maximum ASI value, that is,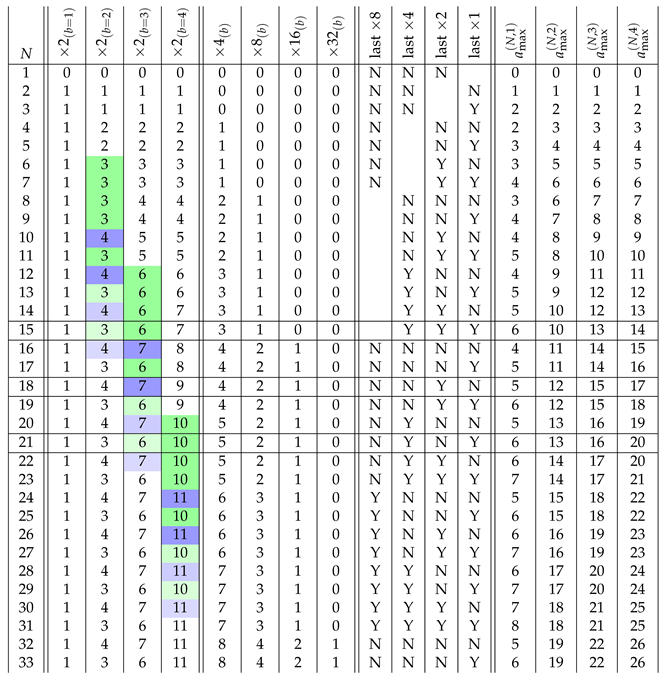 |
7. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
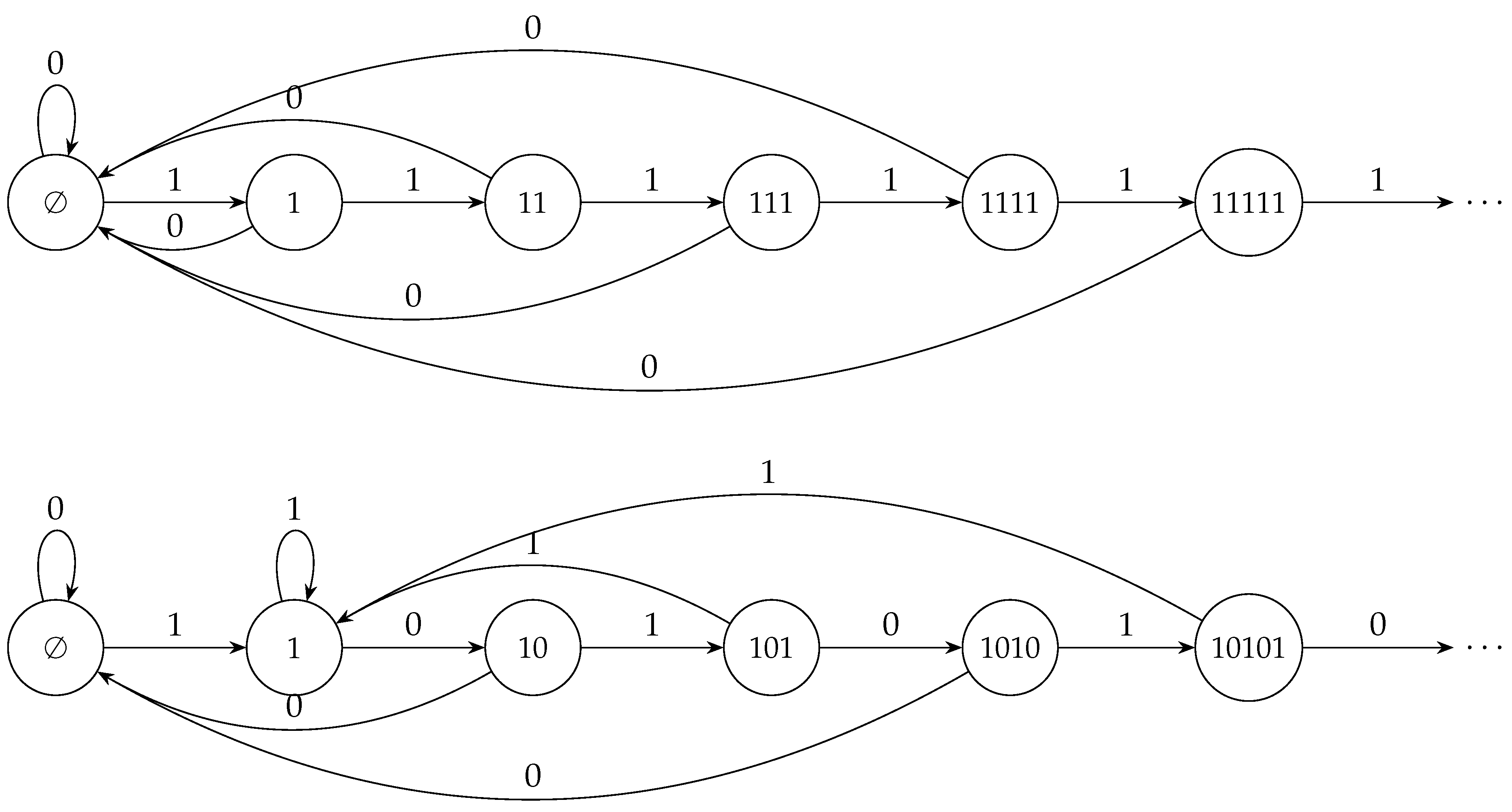
Appendix A. Method A for Generating C (N-1) String

Appendix B. Method B for Generating C (N-1) String
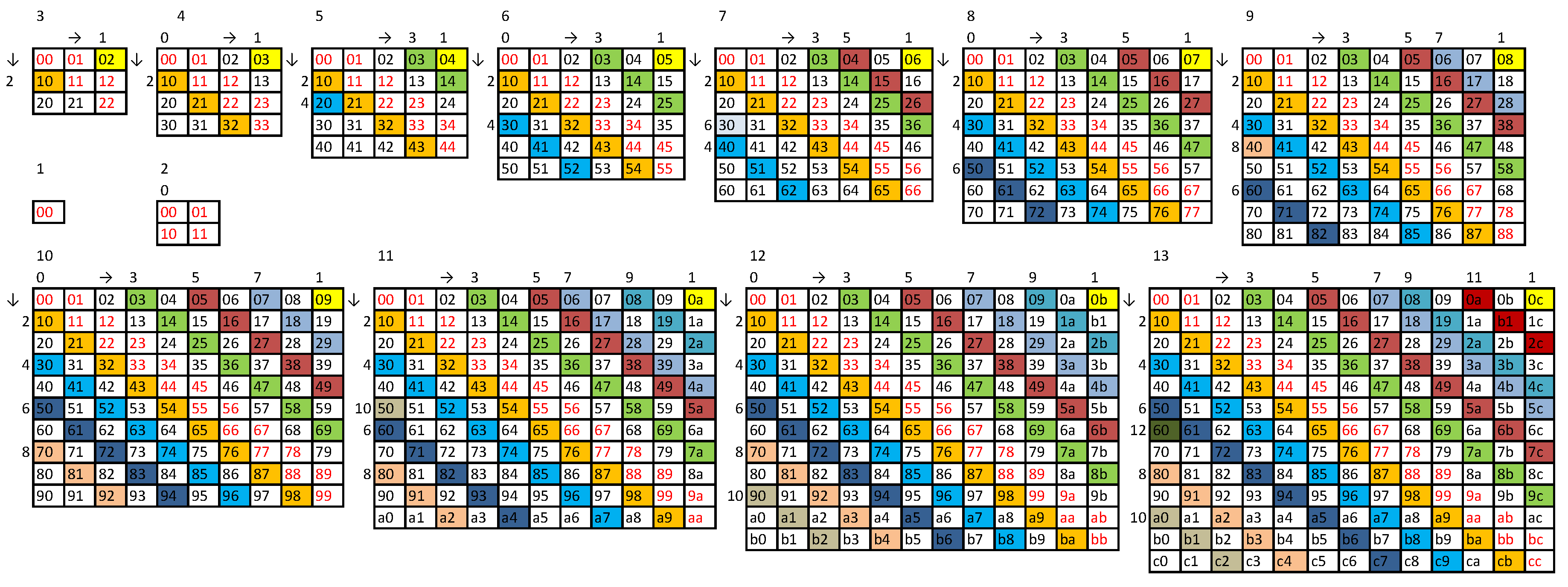
- 1.
- we check subsequent subdiagonals until we find one that does not contain a doublet present in the string formed so far, we append it at the end of this string and proceed to step 2;
- 2.
- we check subsequent superdiagonals until we find one that does not contain a doublet present in the string formed so far, we append it at the end of this string and proceed to step 1.
- Finally, we append 0 if b is even. The method is illustrated in Figure A2 and for generates the strings in the form
Appendix C. A String with Exactly Two Copies of All Doublets and No Repeated Triplets
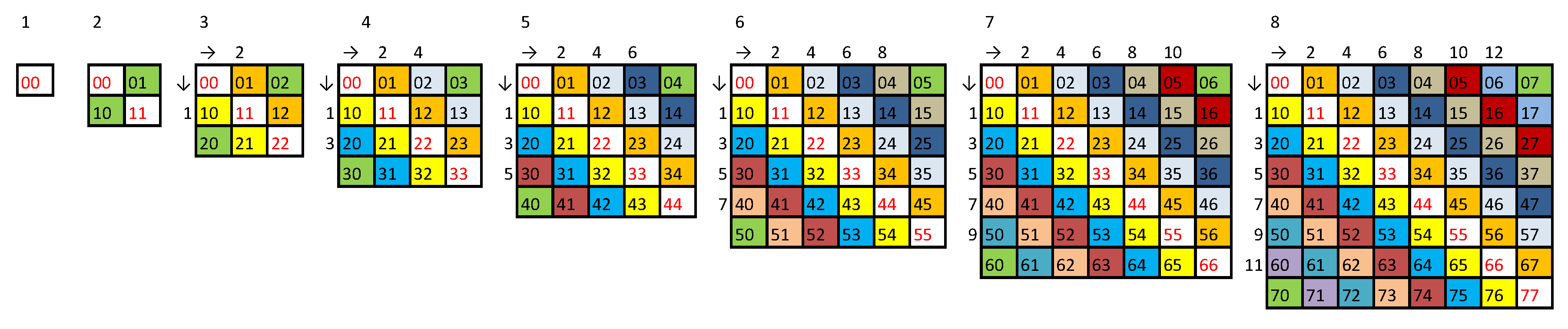
Appendix D. Proof of C (N-1) String Theorem
Appendix E. Proof of C (N-k) String Theorem
Appendix F. Assembly Spaces of Minimum Assembly Index Strings
| N | MIA pathway | MBL pathway (Hamming weight ) | String | ||||
|---|---|---|---|---|---|---|---|
| 2 | 1 | 1 | 1 | 1 | (1) | ||
| 3 | 2 | 2 | 2 | 2 | (1) | ||
| 4 | 2 | 2 | 2 | 2 | (2) | ||
| 5 | 3 | 3 | 3 | 3 | (2) | ||
| 6 | 3 | 3 | 3 | 3 | (3) | ||
| 7 | 3 | 3 | 4 | 4 | (3) | ||
| 8 | 3 | 3 | 3 | 3 | (4) | ||
| 9 | 4 | 4 | 4 | 4 | (4) | ||
| 10 | 4 | 4 | 4 | 4 | (5) | ||
| 11 | 4 | 4 | 5 | 5 | (5) | ||
| 12 | 4 | 4 | 4 | 4 | (6) | ||
| 13 | 4 | 4 | 5 | 5 | (6) | ||
| 14 | 4 | 4 | 5 | 5 | (7) | ||
| 15 | 4 | 5 | 5 | 6 | (6) | ||
| 16 | 4 | 4 | 4 | 4 | (8) | ||
| 17 | 5 | 5 | 5 | 5 | (8) | ||
| 18 | 5 | 5 | 5 | 5 | (9) | ||
| 19 | 5 | 5 | 6 | 6 | (9) | ||
| 20 | 5 | 5 | 5 | 5 | (10) | ||
| 21 | 5 | 5 | 6 | 6 | (10) | ||
| 22 | 5 | 5 | 6 | 6 | (11) | ||
| 23 | 5 | 6 | 6 | 7 | (9) | ||
| 24 | 5 | 5 | 5 | 5 | (12) | ||
| 25 | 5 | 5 | 6 | 6 | (12) | ||
| 26 | 5 | 5 | 6 | 6 | (13) | ||
| 27 | 5 | 6 | 6 | 7 | (12) | ||
| 28 | 5 | 5 | 6 | 6 | (14) | ||
| 29 | 5 | 6 | 7 | 7 | (14) | ||
| 30 | 5 | 6 | 6 | 7 | (15) | ||
| 31 | 5 | 6 | 7 | 8 | (15) | ||
| 32 | 5 | 5 | 5 | 5 | (16) | ||
| 33 | 6 | 6 | 6 | 6 | (16) | ||
| 34 | 6 | 6 | 6 | 6 | (17) | ||
| 35 | 6 | 6 | 7 | 7 | (17) | ||
| 36 | 6 | 6 | 6 | 6 | (18) | ||
| 37 | 6 | 6 | 7 | 7 | (18) | ||
| 38 | 6 | 6 | 7 | 7 | (19) | ||
| 39 | 6 | 6 | 7 | 8 | |||
| 40 | 6 | 6 | 6 | 6 | |||
| 41 | 6 | 6 | 7 | 7 | |||
| 42 | 6 | 6 | 7 | 7 | |||
| 43 | 6 | 7 | 7 | 8 | (17) | ||
| 44 | 6 | 6 | 7 | 7 | (22) | ||
| 45 | 6 | 7 | 7 | 8 | (20) | ||
| 46 | 6 | 7 | 7 | 8 | (23) | ||
| 47 | 6 | 7 | 8 | 9 | (23) | ||
| 48 | 6 | 6 | 6 | 6 | (24) | ||
| 49 | 6 | 7 | 7 | 7 | (24) | ||
| 50 | 6 | 6 | 7 | 7 | (25) | ||
| 51 | 6 | 7 | 7 | 8 | (24) | ||
| 52 | 6 | 6 | 7 | 7 | (26) | ||
| 53 | 6 | 7 | 8 | 8 | (26) | ||
| 54 | 6 | 7 | 7 | 8 | (27) | ||
| 55 | 6 | 7 | 8 | 9 | (27) | ||
| 56 | 6 | 6 | 7 | 7 | (28) | ||
| 57 | 6 | 7 | 8 | 8 | (28) | ||
| 58 | 6 | 7 | 8 | 8 | (29) | ||
| 59 | 6 | 7 | 8 | 9 | (26) | ||
| 60 | 6 | 7 | 7 | 8 | (30) | ||
| 61 | 6 | 8 | 8 | 9 | (30) | ||
| 62 | 6 | 7 | 8 | 9 | (31) | ||
| 63 | 6 | 7 | 8 | 10 | (28) | ||
| 64 | 6 | 6 | 6 | 6 | (32) | ||
| 65 | 7 | 7 | 7 | 7 | (32) | ||
References
- M. Levin, Self-constructing bodies, collective minds: the intersection of cs, cognitive bio, and philosophy (2024).
- M. Levin, Self-Improvising Memory: A Perspective on Memories as Agential, Dynamically Reinterpreting Cognitive Glue, Entropy 26, 481 (2024). [CrossRef]
- D. Hoffman, The case against reality: Why evolution hid the truth from our eyes (WW Norton & Company, 2019).
- D. D. Hoffman and M. Singh, Perception, Evolution, and the Explanatory Scope of Scientific Theories, Journal of Consciousness Studies 31, 29 (2024). [CrossRef]
- Č. Brukner, A No-Go Theorem for Observer-Independent Facts, Entropy 20, (2018). [CrossRef]
- S. Łukaszyk and A. Tomski, Omnidimensional Convex Polytopes, Symmetry 15, (2023). [CrossRef]
- S. Łukaszyk, Shannon entropy of chemical elements, European Journal of Applied Sciences 11, 443–458 (2024). [CrossRef]
- S. Łukaszyk, The Imaginary Universe (on the Three Complementary Sets of Measurement Units Defining Three Dark Electrons) (2024). [CrossRef]
- A. D. Lorenzo, A relation between pythagorean triples and the special theory of relativity (2018). [CrossRef]
- H. Sporn, Pythagorean triples using the relativistic velocity addition formula, The Mathematical Gazette 108, 219 (2024). [CrossRef]
- S. Łukaszyk, Metallic Ratios and Angles of a Real Argument, IPI Letters , 26 (2024). [CrossRef]
- S. M. Marshall, A. R. G. Murray, and L. Cronin, A probabilistic framework for identifying biosignatures using Pathway Complexity, Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences 375, 20160342 (2017). [CrossRef]
- Imari Walker, L. Cronin, A. Drew, S. Domagal-Goldman, T. Fisher, and M. Line, Probabilistic biosignature frameworks, in Planetary Astrobiology, edited by V. Meadows, G. Arney, B. Schmidt, and D. J. Des Marais (University of Arizona Press, 2019) pp. 1–1.
- V. S. Meadows, G. N. Arney, B. E. Schmidt, and D. J. Des Marais, eds., Planetary astrobiology, University of Arizona space science series (The University of Arizona Press ; Houston : Lunar and Planetary Institute, Tucson, 2020) oCLC: 1151198948.
- Y. Liu, C. Mathis, M. D. Bajczyk, S. M. Marshall, L. Wilbraham, and L. Cronin, Exploring and mapping chemical space with molecular assembly trees, Science Advances 7, eabj2465 (2021). [CrossRef]
- S. M. Marshall, C. Mathis, E. Carrick, G. Keenan, G. J. T. Cooper, H. Graham, M. Craven, P. S. Gromski, D. G. Moore, S. I. Walker, and L. Cronin, Identifying molecules as biosignatures with assembly theory and mass spectrometry, Nature Communications 12, 3033 (2021). [CrossRef]
- S. M. Marshall, D. G. Moore, A. R. G. Murray, S. I. Walker, and L. Cronin, Formalising the Pathways to Life Using Assembly Spaces, Entropy 24, 884 (2022). [CrossRef]
- A. Sharma, D. Czégel, M. Lachmann, C. P. Kempes, S. I. Walker, and L. Cronin, Assembly theory explains and quantifies selection and evolution, Nature 622, 321 (2023). [CrossRef]
- M. Jirasek, A. Sharma, J. R. Bame, S. H. M. Mehr, N. Bell, S. M. Marshall, C. Mathis, A. MacLeod, G. J. T. Cooper, M. Swart, R. Mollfulleda, and L. Cronin, Investigating and Quantifying Molecular Complexity Using Assembly Theory and Spectroscopy, ACS Central Science 10, 1054 (2024). [CrossRef]
- S. Łukaszyk and W. Bieniawski, Assembly Theory of Binary Messages, Mathematics 12, 1600 (2024). [CrossRef]
- S. Raubitzek, A. Schatten, P. König, E. Marica, S. Eresheim, and K. Mallinger, Autocatalytic Sets and Assembly Theory: A Toy Model Perspective, Entropy 26, 808 (2024). [CrossRef]
- S. Łukaszyk, On the "Assembly Theory and its Relationship with Computational Complexity" (2024). [CrossRef]
- P. Francis, Dilexit nos: Encyclical letter on the human and divine love of the heart of jesus christ (2024), accessed: 2024-11-01.
- Book of John [1.3] (c90).
- C. P. Kempes, M. Lachmann, A. Iannaccone, G. M. Fricke, M. R. Chowdhury, S. I. Walker, and L. Cronin, Assembly Theory and its Relationship with Computational Complexity (2024). [CrossRef]
- L. Cronin, Exploring assembly index of strings is a good way to show why assembly & entropy are intrinsically different., https://x.com/leecronin/status/1850289225935257665 (2024), accessed: 2024-11-01.
- S. Pagel, A. Sharma, and L. Cronin, Mapping Evolution of Molecules Across Biochemistry with Assembly Theory (2024). [CrossRef]
- S. Łukaszyk, Black Hole Horizons as Patternless Binary Messages and Markers of Dimensionality, in Future Relativity, Gravitation, Cosmology (Nova Science Publishers, 2023) Chap. 15, pp. 317–374. [CrossRef]
- S. Łukaszyk, Life as the explanation of the measurement problem, Journal of Physics: Conference Series 2701, 012124 (2024). [CrossRef]
- G. J. Chaitin, Randomness and Mathematical Proof, Scientific American 232, 47 (1975). [CrossRef]
- A. N. Kolmogorov, Combinatorial foundations of information theory and the calculus of probabilities, Russian Mathematical Surveys 38, 29 (1983). [CrossRef]
- M. Mugur-Schachter, On a Crucial Problem in Probabilities and Solution (2008). [CrossRef]
- M. M. Vopson, The second law of infodynamics and its implications for the simulated universe hypothesis, AIP Advances 13, 105308 (2023). [CrossRef]
- G. Chaitin, Proving Darwin: Making Biology Mathematical (Knopf Doubleday Publishing Group, 2013).
 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





